The domain within your query sequence starts at position 884 and ends at position 1042; the E-value for the AAA domain shown below is 3.08e0.
ERGNALLVGVGGTGKQSLTRLAAHICGYKCMQIELSRGYNYDNFHEDLRKLYKMAGVEDK DMVFLFTDTQIVVEEFLEDINNILNSGEVPNLFEKDELEHVLAATRPKAKEVGISEGNRD EVFQYFISRVRQKLHIVLCMSPVGEAFRSRCRMFPSLVN
AAAATPases associated with a variety of cellular activities |
|---|
| SMART accession number: | SM00382 |
|---|---|
| Description: | AAA - ATPases associated with a variety of cellular activities. This profile/alignment only detects a fraction of this vast family. The poorly conserved N-terminal helix is missing from the alignment. |
| Interpro abstract (IPR003593): | The AAA+ superfamily of ATPases is found in all kingdoms of living organisms where they participate in diverse cellular processes including membrane fusion, proteolysis and DNA replication. Although the terms AAA+ and AAA are often used loosely and interchangeably, the classical AAA family members are distinguished by their possession of the SRH region in the AAA module. Many AAA+ proteins are involved in similar processes to those of AAA proteins (facilitation of protein folding and unfolding, assembly or disassembly of proteins complexes, protein transport and degradation), but others function in replication, recombination, repair and transcription. For a review see [ (PUBMED:11473577) ]. The proteins in this superfamily are characterised by the structural conservation of a central ATPase domain of about 250 amino acids called the AAA+ module. Typically, the AAA+ domain can be divided into two structural subdomains, an N-terminal P-loop NTPase alpha-beta-alpha subdomain that is connected to a smaller C-terminal all-alpha subdomain. The alpha-beta-alpha subdomain adopts a Rossman fold and contains several motifs involved in ATP binding and hydrolysis, including classical motifs Walker A and Walker B [ (PUBMED:11473577) (PUBMED:18466635) ]. The all-alpha subdomain [ (PUBMED:9927482) ], is much less conserved across AAA+ proteins. |
| Family alignment: |
There are 2804801 AAA domains in 2428284 proteins in SMART's nrdb database.
Click on the following links for more information.
- Evolution (species in which this domain is found)
-
Taxonomic distribution of proteins containing AAA domain.
This tree includes only several representative species. The complete taxonomic breakdown of all proteins with AAA domain is also avaliable.
Click on the protein counts, or double click on taxonomic names to display all proteins containing AAA domain in the selected taxonomic class.
- Literature (relevant references for this domain)
-
Primary literature is listed below; Automatically-derived, secondary literature is also avaliable.
- Neuwald AF, Aravind L, Spouge JL, Koonin EV
- AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes.
- Genome Res. 1999; 9: 27-43
- Display abstract
Using a combination of computer methods for iterative database searches and multiple sequence alignment, we show that protein sequences related to the AAA family of ATPases are far more prevalent than reported previously. Among these are regulatory components of Lon and Clp proteases, proteins involved in DNA replication, recombination, and restriction (including subunits of the origin recognition complex, replication factor C proteins, MCM DNA-licensing factors and the bacterial DnaA, RuvB, and McrB proteins), prokaryotic NtrC-related transcription regulators, the Bacillus sporulation protein SpoVJ, Mg2+, and Co2+ chelatases, the Halobacterium GvpN gas vesicle synthesis protein, dynein motor proteins, TorsinA, and Rubisco activase. Alignment of these sequences, in light of the structures of the clamp loader delta' subunit of Escherichia coli DNA polymerase III and the hexamerization component of N-ethylmaleimide-sensitive fusion protein, provides structural and mechanistic insights into these proteins, collectively designated the AAA+ class. Whole-genome analysis indicates that this class is ancient and has undergone considerable functional divergence prior to the emergence of the major divisions of life. These proteins often perform chaperone-like functions that assist in the assembly, operation, or disassembly of protein complexes. The hexameric architecture often associated with this class can provide a hole through which DNA or RNA can be thread; this may be important for assembly or remodeling of DNA-protein complexes.
- Lenzen CU, Steinmann D, Whiteheart SW, Weis WI
- Crystal structure of the hexamerization domain of N-ethylmaleimide-sensitive fusion protein.
- Cell. 1998; 94: 525-36
- Display abstract
N-ethylmaleimide-sensitive fusion protein (NSF) is a cytosolic ATPase required for many intracellular vesicle fusion reactions. NSF consists of an amino-terminal region that interacts with other components of the vesicle trafficking machinery, followed by two homologous ATP-binding cassettes, designated D1 and D2, that possess essential ATPase and hexamerization activities, respectively. The crystal structure of D2 bound to Mg2+-AMPPNP has been determined at 1.75 A resolution. The structure consists of a nucleotide-binding and a helical domain, and it is unexpectedly similar to the first two domains of the clamp-loading subunit delta' of E. coli DNA polymerase III. The structure suggests several regions responsible for coupling of ATP hydrolysis to structural changes in full-length NSF.
- Patel S, Latterich M
- The AAA team: related ATPases with diverse functions.
- Trends Cell Biol. 1998; 8: 65-71
- Display abstract
A new family of related ATPases has emerged, characterized by a highly conserved AAA motif. This motif forms a 230-amino-acid domain that contains Walker homology sequences and imparts ATPase activity. Homology between AAA-family members is confined mostly to the AAA domain, although additional homology outside the AAA motif is present among closely related proteins. AAA proteins act in a variety of cellular functions, including cell-cycle regulation, protein degradation, organelle biogenesis and vesicle-mediated protein transport. The AAA domain is required for protein function, but its exact role and the specific activity that it confers on AAA proteins is still unclear. This review describes current understanding of the AAA protein family.
- Disease (disease genes where sequence variants are found in this domain)
-
SwissProt sequences and OMIM curated human diseases associated with missense mutations within the AAA domain.
Protein Disease Peroxisome biogenesis factor 1 (O43933) (SMART) OMIM:602136: Zellweger syndrome-1
OMIM:214100: Adrenoleukodystrophy, neonatal
OMIM:202370: Refsum disease, infantile
OMIM:266510:ATP-binding cassette sub-family D member 1 (P33897) (SMART) OMIM:300100: Adrenoleukodystrophy ; Adrenomyeloneuropathy Cystic fibrosis transmembrane conductance regulator (P13569) (SMART) OMIM:602421: Cystic fibrosis
OMIM:219700: Congenital bilateral absence of vas deferens
OMIM:277180: Sweat chloride elevation without CF ; {Pancreatitis, idiopathic} ; {Hypertrypsinemia, neonatal}ATP-binding cassette sub-family C member 8 (Q09428) (SMART) OMIM:600509: Persistent hyperinsulinemic hypoglycemia of infancy
OMIM:256450:Spastin (Q9UBP0) (SMART) OMIM:182601: Spastic paraplegia-4
OMIM:604277: Spastic paraplegia-4
OMIM:182601:ATP-binding cassette sub-family A member 1 (O95477) (SMART) OMIM:600046: Tangier disease
OMIM:205400: HDL deficiency, familial
OMIM:604091:Antigen peptide transporter 1 (Q03518) (SMART) OMIM:170260: TRANSPORTER, ATP-BINDING CASSETTE, MAJOR HISTOCOMPATIBILITY COMPLEX DNA repair protein RAD51 homolog 1 (Q06609) (SMART) OMIM:179617: {Breast cancer, susceptibility to}
OMIM:114480:Canalicular multispecific organic anion transporter 1 (Q92887) (SMART) OMIM:601107: Dubin-Johnson syndrome
OMIM:237500:Antigen peptide transporter 2 (Q03519) (SMART) OMIM:170261: Bare lymphocyte syndrome, type I, due to TAP2 deficiency Peroxisome assembly factor 2 (Q13608) (SMART) OMIM:601498: Peroxisomal biogenesis disorder, complementation group 4 Retinal-specific ATP-binding cassette transporter (P78363) (SMART) OMIM:601691: Stargardt disease-1
OMIM:248200: Retinitis pigmentosa-19
OMIM:601718: Cone-rod dystrophy 3 ; Macular dystrophy, age-related, 2
OMIM:153800: Fundus flavimaculatus
OMIM:248200: - Metabolism (metabolic pathways involving proteins which contain this domain)
-
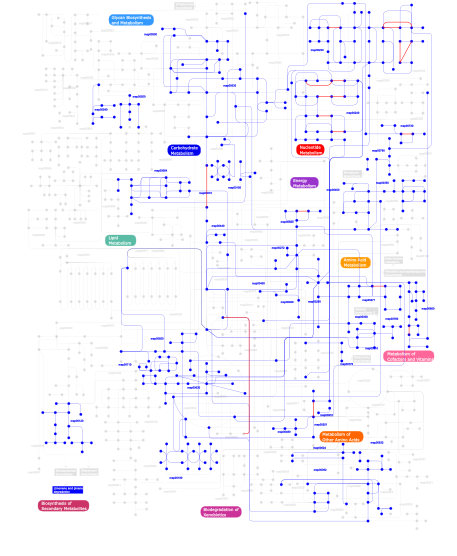
Click the image to view the interactive version of the map in iPath% proteins involved KEGG pathway ID Description 72.32 map02010 ABC transporters - General 4.51 map03060 Protein export 3.93 map03030 DNA replication 3.06 map03090 Type II secretion system 2.69  map00190
map00190Oxidative phosphorylation 1.92  map00195
map00195Photosynthesis 1.85 map02020 Two-component system - General 1.57 map02040 Flagellar assembly 1.26 map03070 Type III secretion system 1.06  map00910
map00910Nitrogen metabolism 0.87 map03050 Proteasome 0.79  map00790
map00790Folate biosynthesis 0.75  map00860
map00860Porphyrin and chlorophyll metabolism 0.67  map00500
map00500Starch and sucrose metabolism 0.57 map04111 Cell cycle - yeast 0.56 map04110 Cell cycle 0.37  map00230
map00230Purine metabolism 0.27 map03080 Type IV secretion system 0.15  map00240
map00240Pyrimidine metabolism 0.14 map05212 Pancreatic cancer 0.11 map04310 Wnt signaling pathway 0.07 map04612 Antigen processing and presentation 0.06  map00400
map00400Phenylalanine, tyrosine and tryptophan biosynthesis 0.06  map00920
map00920Sulfur metabolism 0.06  map00450
map00450Selenoamino acid metabolism 0.05  map00730
map00730Thiamine metabolism 0.05 map04930 Type II diabetes mellitus 0.03  map00780
map00780Biotin metabolism 0.03  map00310
map00310Lysine degradation 0.02 map05120 Epithelial cell signaling in Helicobacter pylori infection 0.02  map00440
map00440Aminophosphonate metabolism 0.02  map00540
map00540Lipopolysaccharide biosynthesis 0.01  map00010
map00010Glycolysis / Gluconeogenesis 0.01  map00271
map00271Methionine metabolism 0.01  map00770
map00770Pantothenate and CoA biosynthesis 0.01  map00260
map00260Glycine, serine and threonine metabolism 0.01  map00350
map00350Tyrosine metabolism 0.01  map00251
map00251Glutamate metabolism 0.01  map00650
map00650Butanoate metabolism 0.01  map00120
map00120Bile acid biosynthesis 0.01  map00710
map00710Carbon fixation 0.01  map00460
map00460Cyanoamino acid metabolism 0.01  map00632
map00632Benzoate degradation via CoA ligation 0.01  map00564
map00564Glycerophospholipid metabolism 0.01  map00300
map00300Lysine biosynthesis 0.01 map00903 Limonene and pinene degradation 0.01  map00624
map006241- and 2-Methylnaphthalene degradation 0.01  map00630
map00630Glyoxylate and dicarboxylate metabolism 0.01  map00480
map00480Glutathione metabolism 0.01  map00550
map00550Peptidoglycan biosynthesis 0.01  map00362
map00362Benzoate degradation via hydroxylation 0.01  map00272
map00272Cysteine metabolism 0.01  map00030
map00030Pentose phosphate pathway This information is based on mapping of SMART genomic protein database to KEGG orthologous groups. Percentage points are related to the number of proteins with AAA domain which could be assigned to a KEGG orthologous group, and not all proteins containing AAA domain. Please note that proteins can be included in multiple pathways, ie. the numbers above will not always add up to 100%.
- Structure (3D structures containing this domain)
3D Structures of AAA domains in PDB
PDB code Main view Title 1b0u 
ATP-BINDING SUBUNIT OF THE HISTIDINE PERMEASE FROM SALMONELLA TYPHIMURIUM 1bmf 
BOVINE MITOCHONDRIAL F1-ATPASE 1cow 
BOVINE MITOCHONDRIAL F1-ATPASE COMPLEXED WITH AUROVERTIN B 1d2n 
D2 DOMAIN OF N-ETHYLMALEIMIDE-SENSITIVE FUSION PROTEIN 1do0 
ORTHORHOMBIC CRYSTAL FORM OF HEAT SHOCK LOCUS U (HSLU) FROM ESCHERICHIA COLI 1do2 
TRIGONAL CRYSTAL FORM OF HEAT SHOCK LOCUS U (HSLU) FROM ESCHERICHIA COLI 1e1q 
BOVINE MITOCHONDRIAL F1-ATPASE AT 100K 1e1r 
BOVINE MITOCHONDRIAL F1-ATPASE INHIBITED BY MG2+ADP AND ALUMINIUM FLUORIDE 1e32 
Structure of the N-Terminal domain and the D1 AAA domain of membrane fusion ATPase p97 1e69 
SMC head domain from Thermotoga maritima 1e79 
Bovine F1-ATPase inhibited by DCCD (dicyclohexylcarbodiimide) 1e94 
HslV-HslU from E.coli 1efr 
BOVINE MITOCHONDRIAL F1-ATPASE COMPLEXED WITH THE PEPTIDE ANTIBIOTIC EFRAPEPTIN 1f3o 
Crystal structure of MJ0796 ATP-binding cassette 1f48 
CRYSTAL STRUCTURE OF THE ESCHERICHIA COLI ARSENITE-TRANSLOCATING ATPASE 1ffh 
N AND GTPASE DOMAINS OF THE SIGNAL SEQUENCE RECOGNITION PROTEIN FFH FROM THERMUS AQUATICUS 1fnn 
CRYSTAL STRUCTURE OF CDC6P FROM PYROBACULUM AEROPHILUM 1fts 
SIGNAL RECOGNITION PARTICLE RECEPTOR FROM E. COLI 1fx0 
Crystal structure of the chloroplast F1-ATPase from spinach 1g18 
RECA-ADP-ALF4 COMPLEX 1g19 
STRUCTURE OF RECA PROTEIN 1g29 
MALK 1g3i 
CRYSTAL STRUCTURE OF THE HSLUV PROTEASE-CHAPERONE COMPLEX 1g41 
CRYSTAL STRUCTURE OF HSLU HAEMOPHILUS INFLUENZAE 1g4a 
CRYSTAL STRUCTURES OF THE HSLVU PEPTIDASE-ATPASE COMPLEX REVEAL AN ATP-DEPENDENT PROTEOLYSIS MECHANISM 1g4b 
CRYSTAL STRUCTURES OF THE HSLVU PEPTIDASE-ATPASE COMPLEX REVEAL AN ATP-DEPENDENT PROTEOLYSIS MECHANISM 1g6h 
CRYSTAL STRUCTURE OF THE ADP CONFORMATION OF MJ1267, AN ATP-BINDING CASSETTE OF AN ABC TRANSPORTER 1g8p 
CRYSTAL STRUCTURE OF BCHI SUBUNIT OF MAGNESIUM CHELATASE 1g8y 
CRYSTAL STRUCTURE OF THE HEXAMERIC REPLICATIVE HELICASE REPA OF PLASMID RSF1010 1g9x 
CHARACTERIZATION OF THE TWINNING STRUCTURE OF MJ1267, AN ATP-BINDING CASSETTE OF AN ABC TRANSPORTER 1gaj 
CRYSTAL STRUCTURE OF A NUCLEOTIDE-FREE ATP-BINDING CASSETTE FROM AN ABC TRANSPORTER 1h8e 
(ADP.AlF4)2(ADP.SO4) bovine F1-ATPase (all three catalytic sites occupied) 1h8h 
Bovine mitochondrial F1-Atpase at 100K 1hqc 
STRUCTURE OF RUVB FROM THERMUS THERMOPHILUS HB8 1hqy 
Nucleotide-Dependent Conformational Changes in a Protease-Associated ATPase HslU 1ht1 
Nucleotide-Dependent Conformational Changes in a Protease-Associated ATPase HslU 1ht2 
Nucleotide-Dependent Conformational Changes in a Protease-Associated ATPase HslU 1ihu 
CRYSTAL STRUCTURE OF THE ESCHERICHIA COLI ARSENITE-TRANSLOCATING ATPASE IN COMPLEX WITH MG-ADP-ALF3 1ii0 
CRYSTAL STRUCTURE OF THE ESCHERICHIA COLI ARSENITE-TRANSLOCATING ATPASE 1ii9 
CRYSTAL STRUCTURE OF THE ESCHERICHIA COLI ARSENITE-TRANSLOCATING ATPASE IN COMPLEX WITH AMP-PNP 1im2 
HslU, Haemophilus Influenzae, Selenomethionine Variant 1in4 
THERMOTOGA MARITIMA RUVB HOLLIDAY JUNCTION BRANCH MIGRATION MOTOR 1in5 
THERMOGOTA MARITIMA RUVB A156S MUTANT 1in6 
THERMOTOGA MARITIMA RUVB K64R MUTANT 1in7 
THERMOTOGA MARITIMA RUVB R170A 1in8 
THERMOTOGA MARITIMA RUVB T158V 1iqp 
Crystal Structure of the Clamp Loader Small Subunit from Pyrococcus furiosus 1ixr 
RuvA-RuvB complex 1ixs 
Structure of RuvB complexed with RuvA domain III 1ixz 
Crystal structure of the FtsH ATPase domain from Thermus thermophilus 1iy0 
Crystal structure of the FtsH ATPase domain with AMP-PNP from Thermus thermophilus 1iy1 
Crystal structure of the FtsH ATPase domain with ADP from Thermus thermophilus 1iy2 
Crystal structure of the FtsH ATPase domain from Thermus thermophilus 1j7k 
THERMOTOGA MARITIMA RUVB P216G MUTANT 1j8m 
Signal Recognition Particle conserved GTPase domain from A. ambivalens 1j8y 
Signal Recognition Particle conserved GTPase domain from A. ambivalens T112A mutant 1jbk 
Crystal Structure of the First Nucelotide Binding Domain of ClpB 1ji0 
Crystal Structure Analysis of the ABC transporter from Thermotoga maritima 1jj7 
Crystal Structure of the C-terminal ATPase domain of human TAP1 1jpj 
GMPPNP Complex of SRP GTPase NG Domain 1jpn 
GMPPNP Complex of SRP GTPase NG Domain 1jr3 
Crystal Structure of the Processivity Clamp Loader Gamma Complex of E. coli DNA Polymerase III 1kmh 
Crystal Structure of spinach chloroplast F1-ATPase complexed with tentoxin 1ksf 
Crystal Structure of ClpA, an HSP100 chaperone and regulator of ClpAP protease: Structural basis of differences in Function of the Two AAA+ ATPase domains 1kyi 
HslUV (H. influenzae)-NLVS Vinyl Sulfone Inhibitor Complex 1l2t 
Dimeric Structure of MJ0796, a Bacterial ABC Transporter Cassette 1l7v 
Bacterial ABC Transporter Involved in B12 Uptake 1l8q 
CRYSTAL STRUCTURE OF DNA REPLICATION INITIATION FACTOR 1ls1 
T. aquaticus Ffh NG Domain at 1.1A Resolution 1lv7 
Crystal Structure of the AAA domain of FtsH 1mab 
RAT LIVER F1-ATPASE 1mo3 
RECA-ADP COMPLEX 1mo4 
RECA-ATP-GAMMA-S COMPLEX 1mo5 
RECA-ATP-GAMMA-S-MG COMPLEX 1mo6 
RECA-DATP-MG COMPLEX 1mt0 
ATP-binding domain of haemolysin B from Escherichia coli 1mv5 
Crystal structure of LmrA ATP-binding domain 1n03 
Model for Active RecA Filament 1n0w 
Crystal structure of a RAD51-BRCA2 BRC repeat complex 1nbm 
THE STRUCTURE OF BOVINE F1-ATPASE COVALENTLY INHIBITED WITH 4-CHLORO-7-NITROBENZOFURAZAN 1ng1 
N AND GTPASE DOMAINS OF THE SIGNAL SEQUENCE RECOGNITION PROTEIN FFH FROM THERMUS AQUATICUS 1njf 
Nucleotide bound form of an isolated E. coli clamp loader gamma subunit 1njg 
Nucleotide-free form of an Isolated E. coli Clamp Loader Gamma Subunit 1nlf 
Crystal Structure of DNA Helicase RepA in complex with sulfate at 1.95 A resolution 1nsf 
D2 HEXAMERIZATION DOMAIN OF N-ETHYLMALEIMIDE SENSITIVE FACTOR (NSF) 1ny5 
Crystal structure of sigm54 activator (AAA+ ATPase) in the inactive state 1ny6 
Crystal structure of sigm54 activator (AAA+ ATPase) in the active state 1o87 
A new MgGDP complex of the Ffh NG domain 1ofh 
Asymmetric complex between HslV and I-domain deleted HslU (H. influenzae) 1ofi 
Asymmetric complex between HslV and I-domain deleted HslU (H. influenzae) 1ohh 
BOVINE MITOCHONDRIAL F1-ATPASE complexed with the inhibitor protein IF1 1ojl 
Crystal structure of a sigma54-activator suggests the mechanism for the conformational switch necessary for sigma54 binding 1okk 
HOMO-HETERODIMERIC COMPLEX OF THE SRP GTPASES 1olo 
Hexameric Replicative DNA Helicase RepA from Plasmid RSF1010 - Cubic Crystal Structure 1oxs 
Crystal structure of GlcV, the ABC-ATPase of the glucose ABC transporter from Sulfolobus solfataricus 1oxt 
Crystal structure of GlcV, the ABC-ATPase of the glucose ABC transporter from Sulfolobus solfataricus 1oxu 
Crystal structure of GlcV, the ABC-ATPase of the glucose ABC transporter from Sulfolobus solfataricus 1oxv 
Crystal structure of GlcV, the ABC-ATPase of the glucose ABC transporter from Sulfolobus solfataricus 1oxx 
Crystal structure of GlcV, the ABC-ATPase of the glucose ABC transporter from Sulfolobus solfataricus 1p9r 
Crystal Structure of Vibrio cholerae putative NTPase EpsE 1p9w 
Crystal Structure of Vibrio cholerae putative NTPase EpsE 1pv4 
X-ray crystal structure of the Rho transcription termination factor in complex with single stranded DNA 1pvo 
X-ray crystal structure of Rho transcription termination factor in complex with ssRNA substrate and ANPPNP 1pzn 
Rad51 (RadA) 1q12 
Crystal Structure of the ATP-bound E. coli MalK 1q1b 
Crystal structure of E. coli MalK in the nucleotide-free form 1q1e 
The ATPase component of E. coli maltose transporter (MalK) in the nucleotide-free form 1q3h 
mouse CFTR NBD1 with AMP.PNP 1qo1 
Molecular Architecture of the Rotary Motor in ATP Synthase from Yeast Mitochondria 1qvr 
Crystal Structure Analysis of ClpB 1qzw 
Crystal structure of the complete core of archaeal SRP and implications for inter-domain communication 1qzx 
Crystal structure of the complete core of archaeal SRP and implications for inter-domain communication 1r0w 
Cystic fibrosis transmembrane conductance regulator (CFTR) nucleotide-binding domain one (NBD1) apo 1r0x 
Cystic fibrosis transmembrane conductance regulator (CFTR) nucleotide-binding domain one (NBD1) with ATP 1r0y 
Cystic fibrosis transmembrane conductance regulator (CFTR) nucleotide-binding domain one (NBD1) with ADP 1r0z 
Phosphorylated Cystic fibrosis transmembrane conductance regulator (CFTR) nucleotide-binding domain one (NBD1) with ATP 1r10 
Cystic fibrosis transmembrane conductance regulator (CFTR) nucleotide-binding domain one (NBD1) with ATP, I4122 space group 1r6b 
High resolution crystal structure of ClpA 1r7r 
The crystal structure of murine p97/VCP at 3.6A 1rea 
STRUCTURE OF THE RECA PROTEIN-ADP COMPLEX 1rj9 
Structure of the heterodimer of the conserved GTPase domains of the Signal Recognition Particle (Ffh) and Its Receptor (FtsY) 1ry1 
Structure of the signal recognition particle interacting with the elongation-arrested ribosome 1s3s 
Crystal structure of AAA ATPase p97/VCP ND1 in complex with p47 C 1sgw 
Putative ABC transporter (ATP-binding protein) from Pyrococcus furiosus Pfu-867808-001 1sky 
CRYSTAL STRUCTURE OF THE NUCLEOTIDE FREE ALPHA3BETA3 SUB-COMPLEX OF F1-ATPASE FROM THE THERMOPHILIC BACILLUS PS3 1sxj 
Crystal Structure of the Eukaryotic Clamp Loader (Replication Factor C, RFC) Bound to the DNA Sliding Clamp (Proliferating Cell Nuclear Antigen, PCNA) 1szp 
A Crystal Structure of the Rad51 Filament 1t4g 
ATPase in complex with AMP-PNP 1u94 
Crystal Structure of E. Coli RecA in a Compressed Helical Filament Form 2 1u98 
""Crystal Structure of E. coli RecA in a Compressed Helical Filament Form3"" 1u99 
""Crystal Structures of E. coli RecA in a Compressed Helical Filament Form 4"" 1ubc 
Structure of Reca Protein 1ube 
MsRecA-ADP Complex 1ubf 
MsREcA-ATPgS complex 1ubg 
MsREcA-dATP complex 1um8 
Crystal structure of helicobacter pylori ClpX 1v43 
Crystal Structure of ATPase subunit of ABC Sugar Transporter 1v5w 
Crystal structure of the human Dmc1 protein 1vci 
Crystal structure of the ATP-binding cassette of multisugar transporter from Pyrococcus horikoshii OT3 complexed with ATP 1vdz 
Crystal structure of A-type ATPase catalytic subunit A from Pyrococcus horikoshii OT3 1vma 
Crystal structure of Cell division protein ftsY (TM0570) from Thermotoga maritima at 1.60 A resolution 1vpl 
Crystal structure of ABC transporter ATP-binding protein (TM0544) from Thermotoga maritima at 2.10 A resolution 1w0j 
Beryllium fluoride inhibited bovine F1-ATPase 1w0k 
Beryllium fluoride inhibited bovine F1-ATPase 1w36 
RecBCD:DNA complex 1xef 
Crystal structure of the ATP/Mg2+ bound composite dimer of HlyB-NBD 1xf9 
Structure of NBD1 from murine CFTR- F508S mutant 1xfa 
Structure of NBD1 from murine CFTR- F508R mutant 1xmi 
Crystal structure of human F508A NBD1 domain with ATP 1xmj 
Crystal structure of human deltaF508 human NBD1 domain with ATP 1xms 
""E. Coli RecA in complex with MnAMP-PNP"" 1xmv 
""E. Coli RecA in complex with MgADP"" 1xp8 
"Deinococcus radiodurans RecA in complex with ATP-gamma-S" 1xpo 
Structural mechanism of inhibition of the Rho transcription termination factor by the antibiotic bicyclomycin 1xpr 
Structural mechanism of inhibition of the Rho transcription termination factor by the antibiotic 5a-formylbicyclomycin (FB) 1xpu 
Structural mechanism of inhibition of the Rho transcription termination factor by the antibiotic 5a-(3-formylphenylsulfanyl)-dihydrobicyclomycin (FPDB) 1xu4 
ATPASE IN COMPLEX WITH AMP-PNP, MAGNESIUM AND POTASSIUM CO-F 1xwi 
Crystal Structure of VPS4B 1xxh 
ATPgS Bound E. Coli Clamp Loader Complex 1xxi 
ADP Bound E. coli Clamp Loader Complex 1ye8 
Crystal Structure of THEP1 from the hyperthermophile Aquifex aeolicus 1yqt 
RNase-L Inhibitor 1yr6 
PAB0955 crystal structure : a GTPase in Apo form from Pyrococcus abyssi 1yr7 
PAB0955 crystal structure : a GTPase in GTP-gamma-S bound form from Pyrococcus abyssi 1yr8 
PAB0955 crystal structure : a GTPase in GTP bound form from Pyrococcus abyssi 1yr9 
PAB0955 crystal structure : a GTPase in GDP and PO4 bound form from Pyrococcus abyssi 1yra 
PAB0955 crystal structure : a GTPase in GDP bound form from Pyrococcus abyssi 1yrb 
PAB0955 crystal structure : a GTPase in GDP and Mg bound form from Pyrococcus abyssi 1yyf 
Correction of X-ray Intensities from an HslV-HslU co-crystal containing lattice translocation defects 1z47 
Structure of the ATPase subunit CysA of the putative sulfate ATP-binding cassette (ABC) transporter from Alicyclobacillus acidocaldarius 1zu4 
Crystal structure of FtsY from Mycoplasma mycoides- space group P21212 1zu5 
Crystal structure of FtsY from Mycoplasma mycoides- space group H32 2awn 
Crystal structure of the ADP-Mg-bound E. Coli MALK (Crystallized with ATP-Mg) 2awo 
Crystal structure of the ADP-Mg-bound E. Coli MALK (Crystallized with ADP-Mg) 2b21 
RADA Recombinase in complex with AMPPNP at pH 6.0 2bbo 
Human NBD1 with Phe508 2bbs 
Human deltaF508 NBD1 with three solubilizing mutations 2bbt 
Human deltaF508 NBD1 with two solublizing mutations. 2bjv 
Crystal Structure of PspF(1-275) R168A mutant 2bjw 
PspF AAA domain 2bke 
Conformational Flexibility Revealed by the Crystal Structure of a Crenarchaeal RadA 2c03 
GDP COMPLEX OF SRP GTPASE FFH NG DOMAIN 2c04 
GMPPCP complex of SRP GTPase Ffh NG Domain at ultra-high resolution 2c96 
Structural basis of the nucleotide driven conformational changes in the AAA domain of transcription activator PspF 2c98 
Structural basis of the nucleotide driven conformational changes in the AAA domain of transcription activator PspF 2c99 
Structural basis of the nucleotide driven conformational changes in the AAA domain of transcription activator PspF 2c9c 
Structural basis of the nucleotide driven conformational changes in the AAA domain of transcription activator PspF 2c9o 
3D Structure of the human RuvB-like helicase RuvBL1 2cbz 
Structure of the human Multidrug Resistance Protein 1 Nucleotide Binding Domain 1 2ce7 
EDTA treated 2cea 
wildtype 2chg 
Replication Factor C domains 1 and 2 2chq 
Replication Factor C ADPNP complex 2chv 
Replication Factor C ADPNP complex 2ck3 
Azide inhibited bovine F1-ATPase 2cnw 
GDPALF4 complex of the SRP GTPases Ffh and FtsY 2d3w 
Crystal Structure of Escherichia coli SufC, an ATPase compenent of the SUF iron-sulfur cluster assembly machinery 2d62 
Crystal structure of multiple sugar binding transport ATP-binding protein 2dfl 
Crystal structure of left-handed RadA filament 2dhr 
Whole cytosolic region of ATP-dependent metalloprotease FtsH (G399L) 2dpy 
Crystal structure of the flagellar type III ATPase FliI 2dr3 
Crystal Structure of RecA superfamily ATPase PH0284 from Pyrococcus horikoshii OT3 2ewv 
Crystal Structure of the Pilus Retraction Motor PilT and Bound ADP 2eww 
Crystal Structure of the Pilus Retraction Motor PilT and Bound ATP 2eyu 
The Crystal Structure of the C-terminal Domain of Aquifex aeolicus PilT 2f1h 
RECOMBINASE IN COMPLEX WITH AMP-PNP and Potassium 2f1i 
Recombinase in Complex with AMP-PNP 2f1j 
Recombinase in Complex with ADP 2f43 
Rat liver F1-ATPase 2ff7 
The ABC-ATPase of the ABC-transporter HlyB in the ADP bound state 2ffa 
Crystal structure of ABC-ATPase H662A of the ABC-transporter HlyB in complex with ADP 2ffb 
The crystal structure of the HlyB-NBD E631Q mutant in complex with ADP 2ffh 
THE SIGNAL SEQUENCE BINDING PROTEIN FFH FROM THERMUS AQUATICUS 2fgj 
Crystal structure of the ABC-cassette H662A mutant of HlyB with bound ATP 2fgk 
Crystal structure of the ABC-cassette E631Q mutant of HlyB with bound ATP 2fpk 
RadA recombinase in complex with ADP 2fpl 
RadA recombinase in complex with AMP-PNP and low concentration of K+ 2fpm 
RadA recombinase in complex with AMP-PNP and high concentration of K+ 2g88 
MSRECA-dATP COMPLEX 2gdj 
Delta-62 RADA recombinase in complex with AMP-PNP and magnesium 2ghi 
Crystal Structure of Plasmodium yoelii Multidrug Resistance Protein 2 2gsz 
Structure of A. aeolicus PilT with 6 monomers per asymmetric unit 2hcb 
Structure of AMPPCP-bound DnaA from Aquifex aeolicus 2hld 
Crystal structure of yeast mitochondrial F1-ATPase 2ht1 
The closed ring structure of the Rho transcription termination factor in complex with nucleic acid in the motor domains 2hyd 
Multidrug ABC transporter SAV1866 2i1q 
RadA Recombinase in complex with Calcium 2ihy 
Structure of the Staphylococcus aureus putative ATPase subunit of an ATP-binding cassette (ABC) transporter 2it1 
Structure of PH0203 protein from Pyrococcus horikoshii 2iw3 
Elongation Factor 3 in complex with ADP 2iwh 
Structure of yeast Elongation Factor 3 in complex with ADPNP 2ix3 
Structure of yeast Elongation Factor 3 2ix8 
Model for eEF3 bound to an 80S ribosome 2ixe 
Crystal structure of the ATPase domain of TAP1 with ATP (D645N mutant) 2ixf 
Crystal structure of the ATPase domain of TAP1 with ATP (D645Q, Q678H mutant) 2ixg 
Crystal structure of the ATPase domain of TAP1 with ATP (S621A, G622V, D645N mutant) 2iy3 
Structure of the E. Coli Signal Regognition Particle 2iyl 
Structure of an FtsY:GDP complex 2j28 
Model of E. coli SRP bound to 70S RNCs 2j37 
Model of Mammalian SRP bound to 80s RNCs 2j45 
Water structure of T. Aquaticus Ffh NG Domain At 1.1A Resolution 2j46 
Water structure of T. Aquaticus Ffh NG Domain At 1.1A Resolution 2j7p 
GMPPNP-stabilized NG domain complex of the SRP GTPases Ffh and FtsY 2jdi 
Ground state structure of F1-ATPase from bovine heart mitochondria ( Bovine F1-ATPase crystallised in the absence of azide) 2jiz 
The Structures of F1-ATPase inhibited by resveratrol, piceatannol and quercetin. 2jj1 
The Structure of F1-ATPase inhibited by piceatannol. 2jj2 
The Structure of F1-ATPase inhibited by quercetin. 2ng1 
N AND GTPASE DOMAINS OF THE SIGNAL SEQUENCE RECOGNITION PROTEIN FFH FROM THERMUS AQUATICUS 2nq2 
An inward-facing conformation of a putative metal-chelate type ABC transporter. 2o5v 
Recombination mediator RecF 2obl 
Structural and biochemical analysis of a prototypical ATPase from the type III secretion system of pathogenic bacteria 2obm 
Structural and biochemical analysis of a prototypical ATPase from the type III secretion system of pathogenic bacteria 2odn 
MSRECA-dATP complex 2odw 
MSrecA-ATP-GAMA-S complex 2oe2 
MSrecA-native-low humidity 95% 2oep 
MSrecA-ADP-complex 2oes 
MSrecA-native-SSB 2ofo 
MSrecA-native 2og2 
Crystal structure of chloroplast FtsY from Arabidopsis thaliana 2olj 
ABC Protein ArtP in complex with ADP/Mg2+ 2olk 
ABC Protein ArtP in complex with ADP-beta-S 2onj 
Structure of the multidrug ABC transporter Sav1866 from S. aureus in complex with AMP-PNP 2onk 
ABC transporter ModBC in complex with its binding protein ModA 2ouk 
ABC Protein ArtP in complex with Sulphate 2oxr 
PAB0955 crystal structure : a GTPase in GDP and Mg bound form from Pyrococcus abyssi (after GTP hydrolysis) 2p65 
Crystal Structure of the first nucleotide binding domain of chaperone ClpB1, putative, (Pv089580) from Plasmodium Vivax 2pcj 
Crystal structure of ABC transporter (aq_297) From Aquifex Aeolicus VF5 2pcl 
Crystal structure of ABC transporter with complex (aq_297) from aquifex aeolicus VF5 2pjz 
The crystal structure of putative Cobalt transport ATP-binding protein (cbiO-2), ST1066 2pmk 
Crystal structures of an isolated ABC-ATPase in complex with TNP-ADP 2pze 
Minimal human CFTR first nucleotide binding domain as a head-to-tail dimer 2pzf 
Minimal human CFTR first nucleotide binding domain as a head-to-tail dimer with delta F508 2pzg 
Minimal human CFTR first nucleotide binding domain as a monomer 2q0h 
ABC Protein ArtP in complex with ADP/Mg2+, ATP-gamma-S hydrolyzed 2q9a 
Structure of Apo FTSY 2q9b 
Structure of FTSY:GMPPNP Complex 2q9c 
Structure of FTSY:GMPPNP with MGCL Complex 2qby 
Crystal structure of a heterodimer of Cdc6/Orc1 initiators bound to origin DNA (from S. solfataricus) 2qe7 
Crystal structure of the f1-atpase from the thermoalkaliphilic bacterium bacillus sp. ta2.a1 2qi9 
ABC-transporter BtuCD in complex with its periplasmic binding protein BtuF 2qp9 
Crystal Structure of S.cerevisiae Vps4 2qpa 
Crystal Structure of S.cerevisiae Vps4 in the presence of ADP 2qy9 
Structure of the NG+1 construct of the E. coli SRP receptor FtsY 2qz4 
Human paraplegin, AAA domain in complex with ADP 2r62 
Crystal structure of Helicobacter pylori ATP dependent protease, FtsH 2r65 
Crystal structure of Helicobacter pylori ATP dependent protease, FtsH ADP complex 2r6a 
Crystal Form BH1 2r6c 
Crystal Form BH2 2r6d 
Crystal Form B1 2r6e 
Crystal Form B2 2r6f 
Crystal Structure of Bacillus stearothermophilus UvrA 2r6g 
The Crystal Structure of the E. coli Maltose Transporter 2reb 
THE STRUCTURE OF THE E. COLI RECA PROTEIN MONOMER AND POLYMER 2rec 
RECA HEXAMER MODEL, ELECTRON MICROSCOPY 2rko 
Crystal Structure of the Vps4p-dimer 2v1u 
STRUCTURE OF THE AEROPYRUM PERNIX ORC1 PROTEIN IN COMPLEX WITH DNA 2v3c 
Crystal structure of the SRP54-SRP19-7S.S SRP RNA complex of M. jannaschii 2v7q 
The structure of F1-ATPase inhibited by I1-60HIS, a monomeric form of the inhibitor protein, IF1. 2vii 
PspF1-275-Mg-AMP 2vye 
Crystal Structure of the DnaC-ssDNA complex 2vyf 
Crystal Structure of the DnaC 2w0m 
Crystal Structure of sso2452 from Sulfolobus solfataricus P2 2w58 
Crystal Structure of the DnaI 2w6e 
Low resolution structures of bovine mitochondrial F1-ATPase during controlled dehydration:hydration state 1. 2w6f 
Low resolution structures of bovine mitochondrial F1-ATPase during controlled dehydration: Hydration State 2. 2w6g 
Low resolution structures of bovine mitochondrial F1-ATPase during controlled dehydration: Hydration State 3. 2w6h 
Low resolution structures of bovine mitochondrial F1-ATPase during controlled dehydration: Hydration State 4A. 2w6i 
Low resolution structures of bovine mitochondrial F1-ATPase during controlled dehydration: Hydration State 4B. 2w6j 
Low resolution structures of bovine mitochondrial F1-ATPase during controlled dehydration: Hydration State 5. 2wpd 
The Mg.ADP inhibited state of the yeast F1c10 ATP synthase 2wss 
The structure of the membrane extrinsic region of bovine ATP synthase 2www 
Crystal Structure of Methylmalonic Acidemia Type A Protein 2x31 
Modelling of the complex between subunits BchI and BchD of magnesium chelatase based on single-particle cryo-EM reconstruction at 7.5 ang 2x8a 
Human Nuclear Valosin containing protein Like (NVL), C-terminal AAA- ATPase domain 2xkv 
Atomic Model of the SRP-FtsY Early Conformation 2xnd 
Crystal structure of bovine F1-c8 sub-complex of ATP Synthase 2xok 
Refined structure of yeast F1c10 ATPase complex to 3 A resolution 2xsz 
The dodecameric human RuvBL1:RuvBL2 complex with truncated domains II 2xxa 
The Crystal Structure of the Signal Recognition Particle (SRP) in Complex with its Receptor(SR) 2xzl 
Upf1-RNA complex 2yhs 
Structure of the E. coli SRP receptor FtsY 2ykg 
Structural insights into RNA recognition by RIG-I 2yyz 
Crystal structure of Sugar ABC transporter, ATP-binding protein 2yz2 
Crystal structure of the ABC transporter in the cobalt transport system 2z43 
Structure of a twinned crystal of RadA 2z4r 
Crystal structure of domain III from the Thermotoga maritima replication initiation protein DnaA 2z4s 
Crystal structure of domain III from the Thermotoga maritima replication initiation protein DnaA 2zam 
Crystal structure of mouse SKD1/VPS4B apo-form 2zan 
Crystal structure of mouse SKD1/VPS4B ATP-form 2zao 
Crystal structure of mouse SKD1/VPS4B ADP-form 2zjb 
Crystal structure of the human Dmc1-M200V polymorphic variant 2zr0 
MSRECA-Q196E mutant 2zr7 
Msreca native form II' 2zr9 
MsRecA Q196E dATP form IV 2zra 
MsRecA Q196E ATPgS 2zrb 
MsRecA Q196E Form II' 2zrc 
MsRecA Q196N Form IV 2zrd 
MsRecA Q196N ADP form IV 2zre 
MsRecA Q196N ATPgS form IV 2zrf 
MsRecA Q196N dATP form IV 2zrg 
MsRecA Q196N dATP form II' 2zrh 
MsRecA Q196A form IV 2zri 
MsRecA Q196A ADP form IV 2zrj 
MsRecA Q196A ATPgS form IV 2zrk 
MsRecA Q196A dATP form IV 2zrl 
MsRecA Q196A dATP FORM II' 2zrm 
MsRecA dATP form IV 2zrn 
MsRecA Form IV 2zro 
MsRecA ADP form IV 2zrp 
MsRecA dATP form II' 2zts 
Crystal structure of KaiC-like protein PH0186 from hyperthermophilic archaea Pyrococcus horikoshii OT3 2zu0 
Crystal structure of SufC-SufD complex involved in the iron-sulfur cluster biosynthesis 2zub 
Left handed RadA 2zuc 
Crystal structure of left-handed RadA filament 2zud 
Crystal Structure of Left-handed RadA Filament 3asy 
ligand-free structure of uridine kinase from thermus thermophilus HB8 3asz 
CMP-complex structure of uridine kinase from Thermus thermophilus HB8 3aux 
Crystal structure of Rad50 bound to ADP 3auy 
Crystal structure of Rad50 bound to ADP 3av0 
Crystal structure of Mre11-Rad50 bound to ATP S 3b5j 
Crystal Structures of the S504A Mutant of an Isolated ABC-ATPase in Complex with TNP-ADP 3b5w 
Crystal Structure of Eschericia coli MsbA 3b5x 
Crystal Structure of MsbA from Vibrio cholerae 3b5y 
Crystal Structure of MsbA from Salmonella typhimurium with AMPPNP 3b5z 
Crystal Structure of MsbA from Salmonella typhimurium with ADP Vanadate 3b60 
Crystal Structure of MsbA from Salmonella typhimurium with AMPPNP, higher resolution form 3b9p 
Spastin 3b9q 
The crystal structure of cpFtsY from Arabidopsis thaliana 3bk7 
Structure of the complete ABCE1/RNAase-L Inhibitor protein from Pyrococcus abysii 3c41 
ABC protein ArtP in complex with AMP-PNP/Mg2+ 3c4j 
ABC protein ArtP in complex with ATP-gamma-S 3cf0 
Structure of D2 subdomain of P97/VCP in complex with ADP 3cf1 
Structure of P97/vcp in complex with ADP/ADP.alfx 3cf2 
Structure of P97/vcp in complex with ADP/AMP-PNP 3cf3 
Structure of P97/vcp in complex with ADP 3cmt 
Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures 3cmu 
Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures 3cmv 
Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures 3cmw 
Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures 3cmx 
Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures 3d31 
ModBC from Methanosarcina acetivorans 3d8b 
Crystal structure of human fidgetin-like protein 1 in complex with ADP 3dhw 
Crystal structure of methionine importer MetNI 3dm5 
Structures of SRP54 and SRP19, the two proteins assembling the ribonucleic core of the Signal Recognition Particle from the archaeon Pyrococcus furiosus. 3dm9 
Structures and Conformations in Solution of the Signal Recognition Particle Receptor from the archaeon Pyrococcus furiosus 3dmd 
Structures and Conformations in Solution of the Signal Recognition Particle Receptor from the archaeon Pyrococcus furiosus 3dzd 
Crystal structure of sigma54 activator NTRC4 in the inactive state 3e1s 
Structure of an N-terminal truncation of Deinococcus radiodurans RecD2 3e70 
Structures and conformations in solution of the Signal Recognition Particle Receptor from the Archaeon Pyrococcus Furiosus 3ec2 
Crystal structure of the DnaC helicase loader 3ecc 
Crystal structure of the DnaC helicase loader in complex with ADP-BeF3 3eie 
Crystal Structure of S.cerevisiae Vps4 in the SO4-bound state 3eih 
Crystal structure of S.cerevisiae Vps4 in the presence of ATPgammaS 3etl 
RadA recombinase from Methanococcus maripaludis in complex with AMPPNP 3ew9 
RADA recombinase from METHANOCOCCUS MARIPALUDIS in complex with AMPPNP and potassium ions 3ewa 
RADA recombinase from METHANOCOCCUS MARIPALUDIS in complex with AMPPNP and ammonium ions 3f9v 
Crystal Structure Of A Near Full-Length Archaeal MCM: Functional Insights For An AAA+ Hexameric Helicase 3fh6 
Crystal structure of the resting state maltose transporter from E. coli 3fks 
Yeast F1 ATPase in the absence of bound nucleotides 3fvq 
Crystal structure of the nucleotide binding domain FbpC complexed with ATP 3fyh 
Recombinase in complex with ADP and metatungstate 3g5u 
Structure of P-glycoprotein Reveals a Molecular Basis for Poly-Specific Drug Binding 3g60 
Structure of P-glycoprotein Reveals a Molecular Basis for Poly-Specific Drug Binding 3g61 
Structure of P-glycoprotein Reveals a Molecular Basis for Poly-Specific Drug Binding 3gd7 
Crystal structure of human NBD2 complexed with N6-Phenylethyl-ATP (P-ATP) 3gfo 
Structure of cbiO1 from clostridium perfringens: Part of the ABC transporter complex cbiONQ. 3glf 
Crystal Structure of the Ecoli Clamp Loader Bound to Primer-Template DNA 3glg 
Crystal Structure of a Mutant (gammaT157A) E. coli Clamp Loader Bound to Primer-Template DNA 3glh 
Crystal Structure of the E. coli clamp loader bound to Psi Peptide 3gli 
Crystal Structure of the E. coli clamp loader bound to Primer-Template DNA and Psi Peptide 3gp8 
Crystal structure of the binary complex of RecD2 with DNA 3gpl 
Crystal structure of the ternary complex of RecD2 with DNA and ADPNP 3h4m 
AAA ATPase domain of the proteasome- activating nucleotidase 3hr8 
Crystal Structure of Thermotoga maritima RecA 3hte 
Crystal structure of nucleotide-free hexameric ClpX 3hu1 
Structure of p97 N-D1 R95G mutant in complex with ATPgS 3hu2 
Structure of p97 N-D1 R86A mutant in complex with ATPgS 3hu3 
Structure of p97 N-D1 R155H mutant in complex with ATPgS 3hws 
Crystal structure of nucleotide-bound hexameric ClpX 3i4l 
Structural characterization for the nucleotide binding ability of subunit A with AMP-PNP of the A1AO ATP synthase 3i72 
Structural characterization for the nucleotide binding ability of subunit A with SO4 of the A1AO ATP synthase 3i73 
Structural characterization for the nucleotide binding ability of subunit A with ADP of the A1AO ATP synthase 3ice 
Rho transcription termination factor bound to RNA and ADP-BeF3 3ikj 
Structural characterization for the nucleotide binding ability of subunit A mutant S238A of the A1AO ATP synthase 3j15 
Model of ribosome-bound archaeal Pelota and ABCE1 3j16 
Models of ribosome-bound Dom34p and Rli1p and their ribosomal binding partners 3j3r 
Structural dynamics of the MecA-ClpC complex revealed by cryo-EM 3j3s 
Structural dynamics of the MecA-ClpC complex revealed by cryo-EM 3j3t 
Structural dynamics of the MecA-ClpC complex revealed by cryo-EM 3j3u 
Structural dynamics of the MecA-ClpC complex revealed by cryo-EM 3j5s 
EttA binds to ribosome exit site and regulates translation by restricting ribosome and tRNA dynamics 3j67 
Structural mechanism of the dynein powerstroke (post-powerstroke state) 3j68 
Structural mechanism of the dynein powerstroke (pre-powerstroke state) 3j94 
3J94 3j95 
3J95 3j96 
3J96 3j97 
3J97 3j98 
3J98 3j99 
3J99 3j9t 
3J9T 3j9u 
3J9U 3j9v 
3J9V 3ja8 
3JA8 3jag 
3JAG 3jah 
3JAH 3jai 
3JAI 3jaj 
3JAJ 3jan 
3JAN 3jc5 
3JC5 3jc6 
3JC6 3jc7 
3JC7 3jc8 
3JC8 3jcm 
3JCM 3jco 
3JCO 3jcp 
3JCP 3jcr 
3JCR 3jvu 
Crystal structure of unliganded P. aeruginosa PilT 3jvv 
Crystal Structure of P. aeruginosa PilT with bound AMP-PCP 3k1j 
Crystal structure of Lon protease from Thermococcus onnurineus NA1 3k70 
Crystal structure of the complete initiation complex of RecBCD 3kds 
apo-FtsH crystal structure 3kl4 
Recognition of a signal peptide by the signal recognition particle 3l0o 
Structure of RNA-free Rho transcription termination factor from Thermotoga maritima 3lda 
Yeast Rad51 H352Y Filament Interface Mutant 3m0e 
Crystal structure of the ATP-bound state of Walker B mutant of NtrC1 ATPase domain 3m4y 
Structural characterization of the subunit A mutant P235A of the A-ATP synthase 3m6a 
Crystal structure of Bacillus subtilis Lon C-terminal domain 3mfy 
Structural characterization of the subunit A mutant F236A of the A-ATP synthase from Pyrococcus horikoshii 3nbx 
Crystal structure of E. coli RavA (Regulatory ATPase variant A) in complex with ADP 3ndb 
Crystal structure of a signal sequence bound to the signal recognition particle 3ng1 
N AND GTPASE DOMAINS OF THE SIGNAL SEQUENCE RECOGNITION PROTEIN FFH FROM THERMUS AQUATICUS 3nh6 
Nucleotide Binding Domain of human ABCB6 (apo structure) 3nh9 
Nucleotide Binding Domain of Human ABCB6 (ATP bound structure) 3nha 
Nucleotide Binding Domain of Human ABCB6 (ADP Mg bound structure) 3nhb 
Nucleotide Binding Domain of Human ABCB6 (ADP bound structure) 3ntu 
RADA RECOMBINASE D302K MUTANT IN COMPLEX with AMP-PNP 3nxs 
Crystal structure of LAO/AO transport system from Mycobacterium smegmatis bound to GDP 3oaa 
Structure of the E.coli F1-ATP synthase inhibited by subunit Epsilon 3oe7 
Structure of four mutant forms of yeast f1 ATPase: gamma-I270T 3oee 
Structure of four mutant forms of yeast F1 ATPase: alpha-F405S 3oeh 
Structure of four mutant forms of yeast F1 ATPase: beta-V279F 3ofn 
Structure of four mutant forms of yeast F1 ATPase: alpha-N67I 3oiy 
Helicase domain of reverse gyrase from Thermotoga maritima 3ozx 
Crystal structure of ABCE1 of Sulfolubus solfataricus (-FeS domain) 3p20 
Crystal structure of vanadate bound subunit A of the A1AO ATP synthase 3p4x 
Helicase domain of reverse gyrase from Thermotoga maritima 3p4y 
Helicase domain of reverse gyrase from Thermotoga maritima - P2 form 3pfi 
2.7 Angstrom resolution crystal structure of a probable holliday junction DNA helicase (ruvB) from Campylobacter jejuni subsp. jejuni NCTC 11168 in complex with adenosine-5'-diphosphate 3pih 
T. maritima UvrA in complex with fluorescein-modified DNA 3puv 
Crystal Structure of an outward-facing MBP-Maltose transporter complex bound to ADP-VO4 3puw 
Crystal Structure of an outward-facing MBP-Maltose transporter complex bound to ADP-AlF4 3pux 
Crystal Structure of an outward-facing MBP-Maltose transporter complex bound to ADP-BeF3 3puy 
Crystal Structure of an outward-facing MBP-Maltose transporter complex bound to AMP-PNP after crystal soaking of the pretranslocation state 3puz 
Crystal Structure of a pre-translocation state MBP-Maltose transporter complex bound to AMP-PNP 3pv0 
Crystal Structure of a pre-translocation state MBP-Maltose transporter complex without nucleotide 3pvs 
Structure and biochemical activities of Escherichia coli MgsA 3pxg 
Structure of MecA121 and ClpC1-485 complex 3pxi 
Structure of MecA108:ClpC 3qf4 
Crystal structure of a heterodimeric ABC transporter in its inward-facing conformation 3qf7 
The Mre11:Rad50 complex forms an ATP dependent molecular clamp in DNA double-strand break repair 3qg5 
The Mre11:Rad50 complex forms an ATP dependent molecular clamp in DNA double-strand break repair 3qjy 
Crystal structure of P-loop G234A mutant of subunit A of the A1AO ATP synthase 3qkt 
Rad50 ABC-ATPase with adjacent coiled-coil region in complex with AMP-PNP 3qku 
Mre11 Rad50 binding domain in complex with Rad50 and AMP-PNP 3qmz 
Crystal structure of the cytoplasmic dynein heavy chain motor domain 3r8f 
Protein-DNA complex 3rlf 
Crystal structure of the maltose-binding protein/maltose transporter complex in an outward-facing conformation bound to MgAMPPNP 3sdz 
Structural characterization of the subunit A mutant F427W of the A-ATP synthase from Pyrococcus horikoshii 3se0 
Structural characterization of the subunit A mutant F508W of the A-ATP synthase from Pyrococcus horikoshii 3si7 
The crystal structure of the NBD1 domain of the mouse CFTR protein, deltaF508 mutant 3syk 
Crystal structure of the AAA+ protein CbbX, selenomethionine structure 3syl 
Crystal structure of the AAA+ protein CbbX, native structure 3tif 
Dimeric structure of a post-hydrolysis state of the ATP-binding cassette MJ0796 bound to ADP and Pi 3tk1 
Crystal structure of a MeaB and Rv1496 ortholog from Mycobacterium thermoresistible bound to GDP 3tui 
Inward facing conformations of the MetNI methionine ABC transporter: CY5 native crystal form 3tuj 
Inward facing conformations of the MetNI methionine ABC transporter: DM crystal form 3tuz 
Inward facing conformations of the MetNI methionine ABC transporter: CY5 SeMet soak crystal form 3u5z 
Structure of T4 Bacteriophage clamp loader bound to the T4 clamp, primer-template DNA, and ATP analog 3u60 
Structure of T4 Bacteriophage Clamp Loader Bound To Open Clamp, DNA and ATP Analog 3u61 
Structure of T4 Bacteriophage Clamp Loader Bound To Closed Clamp, DNA and ATP Analog and ADP 3uk6 
Crystal Structure of the Tip48 (Tip49b) hexamer 3upu 
Crystal structure of the T4 Phage SF1B Helicase Dda 3uwx 
Crystal structure of UvrA-UvrB complex 3ux8 
Crystal structure of UvrA 3vfd 
Human spastin AAA domain 3vkg 
X-ray structure of an MTBD truncation mutant of dynein motor domain 3vkh 
X-ray structure of a functional full-length dynein motor domain 3vx4 
Crystal Structure of the Nucleotide-Binding Domain of S. mutans ComA, a Bifunctional ATP-binding Cassette Transporter Involved in the Quorum-sensing Pathway 3w34 
Ternary complex of Thermus thermophilus HB8 uridine-cytidine kinase with substrates 3w8r 
Mutant structure of Thermus thermophilus HB8 uridine-cytidine kinase 3whk 
Crystal structure of PAN-Rpt5C chimera 3whl 
Crystal structure of Nas2 N-terminal domain complexed with PAN-Rpt5C chimera 3wme 
Crystal structure of an inward-facing eukaryotic ABC multidrug transporter 3wmf 
Crystal structure of an inward-facing eukaryotic ABC multitrug transporter G277V/A278V/A279V mutant 3wmg 
Crystal structure of an inward-facing eukaryotic ABC multidrug transporter G277V/A278V/A279V mutant in complex with an cyclic peptide inhibitor, aCAP 3zd6 
Snapshot 1 of RIG-I scanning on RNA duplex 3zd7 
Snapshot 3 of RIG-I scanning on RNA duplex 3zdq 
STRUCTURE OF THE HUMAN MITOCHONDRIAL ABC TRANSPORTER, ABCB10 ( NUCLEOTIDE-FREE FORM) 3zgx 
Crystal structure of the kleisin-N SMC interface in prokaryotic condensin 3zia 
The structure of F1-ATPase from Saccharomyces cerevisiae inhibited by its regulatory protein IF1 3zn8 
Structural Basis of Signal Sequence Surveillance and Selection by the SRP-SR Complex 3zry 
Rotor architecture in the F(1)-c(10)-ring complex of the yeast F-ATP synthase 3zuh 
Negative stain EM Map of the AAA protein CbbX, a red-type Rubisco activase from R. sphaeroides 4a6p 
RadA C-terminal ATPase domain from Pyrococcus furiosus 4a6x 
RadA C-terminal ATPase domain from Pyrococcus furiosus bound to ATP 4a82 
Fitted model of staphylococcus aureus sav1866 model ABC transporter in the human cystic fibrosis transmembrane conductance regulator volume map EMD-1966. 4ag5 
Structure of VirB4 of Thermoanaerobacter pseudethanolicus 4ag6 
Structure of VirB4 of Thermoanaerobacter pseudethanolicus 4ai6 
Dynein Motor Domain - ADP complex 4ak9 
Structure of chloroplast FtsY from Physcomitrella patens 4akg 
Dynein Motor Domain - ATP complex 4akh 
Dynein Motor Domain - AMPPNP complex 4aki 
Dynein Motor Domain - LuAc derivative 4asu 
F1-ATPase in which all three catalytic sites contain bound nucleotide, with magnesium ion released in the Empty site 4ay2 
Capturing 5' tri-phosphorylated RNA duplex by RIG-I 4ayt 
STRUCTURE OF THE HUMAN MITOCHONDRIAL ABC TRANSPORTER, ABCB10 4ayw 
STRUCTURE OF THE HUMAN MITOCHONDRIAL ABC TRANSPORTER, ABCB10 (PLATE FORM) 4ayx 
STRUCTURE OF THE HUMAN MITOCHONDRIAL ABC TRANSPORTER, ABCB10 (ROD FORM B) 4b2i 
Humanised monomeric RadA in complex with indazole 4b2l 
Humanised monomeric RadA in complex with L-methylester tryptophan 4b2p 
RadA C-terminal ATPase domain from Pyrococcus furiosus bound to GTP 4b2q 
Model of the yeast F1Fo-ATP synthase dimer based on subtomogram average 4b32 
Humanised monomeric RadA in complex with napht-1-ol 4b33 
Humanised monomeric RadA in complex with napht-2-ol 4b34 
Humanised monomeric RadA in complex with 2-amino benzothiazole 4b35 
Humanised monomeric RadA in complex with 4-methylester indole 4b3b 
Humanised monomeric RadA in complex with FHTA tetrapeptide 4b3c 
Humanised monomeric RadA in complex with 5-hydroxy indole 4b3d 
Humanised monomeric RadA in complex with 5-methyl indole 4b3f 
crystal structure of Ighmbp2 helicase 4b3g 
crystal structure of Ighmbp2 helicase in complex with RNA 4bgd 
Crystal structure of Brr2 in complex with the Jab1/MPN domain of Prp8 4bpb 
STRUCTURAL INSIGHTS INTO RNA RECOGNITION BY RIG-I 4bs1 
MuB is an AAAplus ATPase that forms helical filaments to control target selection for DNA transposition 4bt0 
MuB is an AAAplus ATPase that forms helical filaments to control target selection for DNA transposition 4bt1 
MuB is an AAAplus ATPase that forms helical filaments to control target selection for DNA transposition 4c3z 
4C3Z 4c7o 
The structural basis of FtsY recruitment and GTPase activation by SRP RNA 4ciu 
Crystal structure of E. coli ClpB 4cr2 
Deep classification of a large cryo-EM dataset defines the conformational landscape of the 26S proteasome 4cr3 
Deep classification of a large cryo-EM dataset defines the conformational landscape of the 26S proteasome 4cr4 
Deep classification of a large cryo-EM dataset defines the conformational landscape of the 26S proteasome 4crm 
4CRM 4d2q 
4D2Q 4d2u 
4D2U 4d2x 
4D2X 4d6p 
4D6P 4d80 
4D80 4d81 
4D81 4d82 
4D82 4dbl 
Crystal structure of E159Q mutant of BtuCDF 4dc9 
Hexameric ring of Methanococcus voltae RadA 4ddt 
Thermotoga maritima reverse gyrase, C2 FORM 2 4ddu 
Thermotoga maritima reverse gyrase, C2 FORM 1 4ddv 
Thermotoga maritima reverse gyrase, triclinic form 4ddw 
Thermotoga maritima reverse gyrase, c-centered orthorhombic form 4ddx 
Thermotoga maritima reverse gyrase, primitive monoclinic form 4eiw 
Whole cytosolic region of atp-dependent metalloprotease FtsH (G399L) 4esv 
A New Twist on the Translocation Mechanism of Helicases from the Structure of DnaB with its Substrates 4f4c 
The Crystal Structure of the Multi-Drug Transporter 4f91 
Brr2 Helicase Region 4f92 
Brr2 Helicase Region S1087L 4f93 
Brr2 Helicase Region S1087L, Mg-ATP 4fct 
Crystal structure of the C-terminal domain of ClpB 4fcv 
Crystal structure of the C-terminal domain of ClpB 4fcw 
Crystal structure of the C-terminal domain of ClpB 4fd2 
Crystal structure of the C-terminal domain of ClpB 4fdg 
Crystal Structure of an Archaeal MCM Filament 4fi3 
Structure of vitamin B12 transporter BtuCD-F in a nucleotide-bound state 4fin 
Crystal Structure of EttA (formerly YjjK) - an E. coli ABC-type ATPase 4fwi 
Crystal structure of the nucleotide-binding domain of a dipeptide ABC transporter 4g1u 
X-ray structure of the bacterial heme transporter HmuUV from Yersinia pestis 4hlu 
Structure of the EcfA-A' heterodimer bound to ADP 4hse 
Crystal structure of ClpB NBD1 in complex with guanidinium chloride and ADP 4huq 
Crystal Structure of a transporter 4hyy 
Filament of octameric rings of DMC1 recombinase from Homo sapiens 4hzi 
Crystal structure of the Leptospira interrogans ATPase subunit of an orphan ABC transporter 4hzu 
Structure of a bacterial energy-coupling factor transporter 4i34 
Crystal Structure of W-W-W ClpX Hexamer 4i4l 
Crystal Structure of Nucleotide-Bound W-W-W ClpX Hexamer 4i5o 
Crystal Structure of W-W-R ClpX Hexamer 4i63 
Crystal Structure of E-R ClpX Hexamer 4i81 
Crystal Structure of ATPgS bound ClpX Hexamer 4i99 
Crystal structure of the SmcHead bound to the C-winged helix domain of ScpA 4i9k 
Crystal structure of symmetric W-W-W ClpX Hexamer 4jbw 
Crystal structure of E. coli maltose transporter MalFGK2 in complex with its regulatory protein EIIAglc 4k8o 
4K8O 4khz 
Crystal structure of the maltose-binding protein/maltose transporter complex in an pre-translocation conformation bound to maltoheptaose 4ki0 
Crystal structure of the maltose-binding protein/maltose transporter complex in an outward-facing conformation bound to maltohexaose 4kit 
Crystal structure of human Brr2 in complex with the Prp8 Jab1/MPN domain 4kln 
Structure of p97 N-D1 A232E mutant in complex with ATPgS 4ko8 
Structure of p97 N-D1 R155H mutant in complex with ATPgS 4kod 
Structure of p97 N-D1 R155H mutant in complex with ADP 4ksb 
Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain 4ksc 
Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain 4ksd 
Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain 4ksr 
Crystal Structure of the Vibrio cholerae ATPase GspE Hexamer 4kss 
Crystal Structure of Vibrio cholerae ATPase GspsE Hexamer 4l15 
Crystal structure of FIGL-1 AAA domain 4l16 
Crystal structure of FIGL-1 AAA domain in complex with ADP 4l4u 
Crystal structure of construct containing A. aeolicus NtrC1 receiver, central and DNA binding domains 4lcb 
Structure of Vps4 homolog from Acidianus hospitalis 4lgm 
Crystal Structure of Sulfolobus solfataricus Vps4 4lj4 
ClpB NBD2 from T. thermophilus, nucleotide-free 4lj5 
ClpB NBD2 from T. thermophilus in complex with ADP 4lj6 
ClpB NBD2 from T. thermophilus in complex with AMPPCP 4lj7 
ClpB NBD2 K601Q from T. thermophilus in complex with MANT-dADP 4lj8 
ClpB NBD2 R621Q from T. thermophilus in complex with ADP 4lj9 
ClpB NBD2 R621Q from T. thermophilus in complex with AMPPCP 4lja 
ClpB NBD2 R621Q from T. thermophilus in complex with AMPPCP and guanidinium chloride 4lsg 
Structure of Mouse P-Glycoprotein 4ly6 
Nucleotide-induced asymmetry within ATPase activator ring drives s54-RNAP interaction and ATP hydrolysis 4lzz 
Nucleotide-induced asymmetry within atpase activator ring drives s54-RNAP interaction and ATP hydrolysis 4m1m 
Corrected Structure of Mouse P-glycoprotein 4m2s 
Corrected Structure of Mouse P-glycoprotein bound to QZ59-RRR 4m2t 
Corrected Structure of Mouse P-glycoprotein bound to QZ59-SSS 4m4w 
Mechanistic implications for the bacterial primosome assembly of the structure of a helicase-helicase loader complex 4mki 
Cobalt transporter ATP-binding subunit 4mrn 
Structure of a bacterial Atm1-family ABC transporter 4mrp 
Structure of a bacterial Atm1-family ABC transporter 4mrr 
Structure of a bacterial Atm1-family ABC transporter 4mrs 
Structure of a bacterial Atm1-family ABC transporter 4mrv 
Structure of a bacterial Atm1-family ABC transporter 4myc 
Structure of the mitochondrial ABC transporter, Atm1 4myh 
Structure of the Glutathione bound mitochondrial ABC transporter, Atm1 4nch 
Crystal Structure of Pyrococcus furiosis Rad50 L802W mutation 4nci 
Crystal Structure of Pyrococcus furiosis Rad50 R805E mutation 4ncj 
Crystal Structure of Pyrococcus furiosis Rad50 R805E mutation with ADP Beryllium Flouride 4nck 
Crystal Structure of Pyrococcus furiosis Rad50 R797G mutation 4nh0 
4NH0 4nmn 
Aquifex aeolicus replicative helicase (DnaB) complexed with ADP, at 3.3 resolution 4nph 
4NPH 4on9 
4ON9 4oqf 
4OQF 4p31 
Crystal structure of a selenomethionine derivative of E. coli LptB in complex with ADP-Magensium 4p32 
Crystal structure of E. coli LptB in complex with ADP-magnesium 4p33 
Crystal structure of E. coli LptB-E163Q in complex with ATP-sodium 4pht 
4PHT 4pl0 
4PL0 4po1 
4PO1 4po8 
4PO8 4po9 
4PO9 4poa 
4POA 4ppf 
4PPF 4ppg 
4PPG 4ppn 
4PPN 4ppq 
4PPQ 4pqf 
4PQF 4pqr 
4PQR 4pqy 
4PQY 4pr0 
4PR0 4psa 
4PSA 4psk 
4PSK 4psv 
4PSV 4ptl 
4PTL 4q4a 
4Q4A 4q4h 
4Q4H 4q4j 
4Q4J 4q4l 
Crystal structure of an ATP synthase subunit beta 1 (F1-B1) from Burkholderia thailandensis 4q7k 
4Q7K 4q7l 
4Q7L 4q7m 
4Q7M 4q9h 
4Q9H 4q9i 
4Q9I 4q9j 
4Q9J 4q9k 
4Q9K 4q9l 
4Q9L 4qc2 
4QC2 4qhs 
4QHS 4qht 
4QHT 4qkq 
4QKQ 4qnm 
4QNM 4qnr 
4QNR 4qos 
4QOS 4r7y 
4R7Y 4r9u 
4R9U 4rfs 
4RFS 4rh7 
4RH7 4rvc 
4RVC 4ry2 
4RY2 4s0f 
4S0F 4tqu 
4TQU 4tqv 
4TQV 4tsf 
4TSF 4tt3 
4TT3 4twz 
4TWZ 4u00 
4U00 4u02 
4U02 4ue5 
4UE5 4upb 
4UPB 4uqo 
4UQO 4w8f 
4W8F 4w9m 
4W9M 4wbs 
4WBS 4wia 
4WIA 4wvy 
4WVY 4ww0 
4WW0 4ww4 
4WW4 4wz6 
4WZ6 4xd7 
4XD7 4xgc 
4XGC 4xgu 
4XGU 4xig 
4XIG 4xtc 
4XTC 4xwk 
4XWK 4yer 
4YER 4yms 
4YMS 4ymt 
4YMT 4ymu 
4YMU 4ymv 
4YMV 4ymw 
4YMW 4ypl 
4YPL 4ypn 
4YPN 4yxw 
4YXW 4z1m 
4Z1M 4z8x 
4Z8X 4zir 
4ZIR 4zpx 
4ZPX 5a5b 
5A5B 5ara 
5ARA 5are 
5ARE 5arh 
5ARH 5ari 
5ARI 5awf 
5AWF 5awg 
5AWG 5b0o 
5B0O 5b57 
5B57 5b58 
5B58 5bq5 
5BQ5 5bw9 
5BW9 5c18 
5C18 5c19 
5C19 5c1a 
5C1A 5c1b 
5C1B 5c73 
5C73 5c76 
5C76 5c78 
5C78 5cdf 
5CDF 5d3m 
5D3M 5d80 
5D80 5da9 
5DA9 5dca 
5DCA 5dgx 
5DGX 5dn6 
5DN6 5dny 
5DNY 5do7 
5DO7 5dyg 
5DYG 5dyi 
5DYI 5e3h 
5E3H 5e7p 
5E7P 5eg1 
5EG1 5ep0 
5EP0 5ep1 
5EP1 5ep2 
5EP2 5ep3 
5EP3 5ep4 
5EP4 5eqt 
5EQT 5eum 
5EUM 5exp 
5EXP 5exs 
5EXS 5ext 
5EXT 5exx 
5EXX 5f3w 
5F3W 5f98 
5F98 5f9f 
5F9F 5f9h 
5F9H 5fij 
5FIJ 5fik 
5FIK 5fil 
5FIL 5fl3 
5FL3 5fl7 
5FL7 5fl8 
5FL8 5fm6 
5FM6 5fm7 
5FM7 5fos 
5FOS 5fot 
5FOT 5fou 
5FOU 5fov 
5FOV 5fow 
5FOW 5fox 
5FOX 5fpk 
5FPK 5ftb 
5FTB 5ftc 
5FTC 5ftd 
5FTD 5fte 
5FTE 5ftf 
5FTF 5ftj 
5FTJ 5ftk 
5FTK 5ftl 
5FTL 5ftm 
5FTM 5ftn 
5FTN 5g4f 
5G4F 5g4g 
5G4G 5gad 
5GAD 5gaf 
5GAF 5gag 
5GAG 5gah 
5GAH 5gan 
5GAN 5gao 
5GAO 5gap 
5GAP 5gjq 
5GJQ 5gjr 
5GJR 5gm6 
5GM6 5hkk 
5HKK 5idv 
5IDV 5ifs 
5IFS 5ifw 
5IFW 5ik2 
5IK2 5j4h 
5J4H 5j4k 
5J4K 5j4l 
5J4L 5jcs 
5JCS 5jec 
5JEC 5jed 
5JED 5jee 
5JEE 5jfg 
5JFG 5ji2 
5JI2 5jji 
5JJI 5jjk 
5JJK 5jjl 
5JJL 5jrj 
5JRJ 5jsz 
5JSZ 5jzc 
5JZC 5kdd 
5KDD 5kne 
5KNE 5ko2 
5KO2 5kpd 
5KPD 5kpi 
5KPI 5kpj 
5KPJ 5l3q 
5L3Q 5l3r 
5L3R 5l3s 
5L3S 5l3v 
5L3V 5l3w 
5L3W 5l4g 
5L4G 5l8v 
5L8V 5lb2 
5LB2 5lb4 
5LB4 5lbi 
5LBI 5ld2 
5LD2 5lj5 
5LJ5 5lqw 
5LQW 5lqx 
5LQX 5lqy 
5LQY 5lqz 
5LQZ 5lw7 
5LW7 5lzv 
5LZV 5swj 
5SWJ 5swl 
5SWL 5syp 
5SYP 5syr 
5SYR 5t0c 
5T0C 5t0g 
5T0G 5t0h 
5T0H 5t0i 
5T0I 5t0j 
5T0J - Links (links to other resources describing this domain)
-
PROSITE AAA INTERPRO IPR003593 PFAM AAA

