The domain within your query sequence starts at position 1218 and ends at position 1303; the E-value for the B41 domain shown below is 3e-45.
VECKEGGFPQELWLGVSADAVSVYKRGEGKPLEVFQYEHILSFGAPLANTYKIVVDEREL LFETSEVVDVAKLMKAYISMIVKKRY
The domain was found using the schnipsel database
B41Band 4.1 homologues |
|---|
| SMART accession number: | SM00295 |
|---|---|
| Description: | Also known as ezrin/radixin/moesin (ERM) protein domains. Present in myosins, ezrin, radixin, moesin, protein tyrosine phosphatases. Plasma membrane-binding domain. These proteins play structural and regulatory roles in the assembly and stabilization of specialized plasmamembrane domains. Some PDZ domain containing proteins bind one or more of this family. Now includes JAKs. |
| Interpro abstract (IPR019749): | The FERM domain (F for 4.1 protein, E for ezrin, R for radixin and M for moesin) is a widespread protein module involved in localising proteins to the plasma membrane [ (PUBMED:9757824) ]. FERM domains are found in a number of cytoskeletal-associated proteins that associate with various proteins at the interface between the plasma membrane and the cytoskeleton. The FERM domain is located at the N terminus of the majority of FERM-containing proteins [ (PUBMED:9757824) (PUBMED:10847681) ], which includes:
Ezrin, moesin, and radixin are highly related proteins (ERM protein family), but the other proteins in which the FERM domain is found do not share any region of similarity outside of this domain. ERM proteins are made of three domains, the FERM domain, a central helical domain and a C-terminal tail domain, which binds F-actin. The amino-acid sequence of the FERM domain is highly conserved among ERM proteins and is responsible for membrane association by direct binding to the cytoplasmic domain or tail of integral membrane proteins. ERM proteins are regulated by an intramolecular association of the FERM and C-terminal tail domains that masks their binding sites for other molecules. For cytoskeleton-membrane cross-linking, the dormant molecules becomes activated and the FERM domain attaches to the membrane by binding specific membrane proteins, while the last 34 residues of the tail bind actin filaments. Aside from binding to membranes, the activated FERM domain of ERM proteins can also bind the guanine nucleotide dissociation inhibitor of Rho GTPase (RhoDGI), which suggests that in addition to functioning as a cross-linker, ERM proteins may influence Rho signalling pathways. The crystal structure of the FERM domain reveals that it is composed of three structural modules (F1, F2, and F3) that together form a compact clover-shaped structure [ (PUBMED:10970839) ]. The FERM domain has also been called the amino-terminal domain, the 30kDa domain, 4.1N30, the membrane-cytoskeletal-linking domain, the ERM-like domain, the ezrin-like domain of the band 4.1 superfamily, the conserved N-terminal region, and the membrane attachment domain [ (PUBMED:9757824) ]. |
| Family alignment: |
There are 33941 B41 domains in 32529 proteins in SMART's nrdb database.
Click on the following links for more information.
- Evolution (species in which this domain is found)
-
Taxonomic distribution of proteins containing B41 domain.
This tree includes only several representative species. The complete taxonomic breakdown of all proteins with B41 domain is also avaliable.
Click on the protein counts, or double click on taxonomic names to display all proteins containing B41 domain in the selected taxonomic class.
- Cellular role (predicted cellular role)
-
Cellular role: signalling
Binding / catalysis: Plasma-membrane-binding - Literature (relevant references for this domain)
-
Primary literature is listed below; Automatically-derived, secondary literature is also avaliable.
- Doi Y et al.
- Normal development of mice and unimpaired cell adhesion/cell motility/actin-based cytoskeleton without compensatory up-regulation of ezrin or radixin in moesin gene knockout.
- J Biol Chem. 1999; 274: 2315-21
- Display abstract
Ezrin/radixin/moesin (ERM) proteins are general cross-linkers between the plasma membrane and actin filaments. Because their expression is regulated in a tissue-specific manner, each ERM protein has been proposed to have unique functions. On the other hand, experiments at the cellular level and in vitro have suggested their functional redundancy. To assess the possible unique functions of ERM proteins in vivo, the moesin gene located on the X chromosome was disrupted by gene targeting in embryonic stem cells. Male mice hemizygous for the mutation as well as homozygous females were completely devoid of moesin but developed normally and were fertile, with no obvious histological abnormalities in any of the tissues examined. In the tissues of the mutant mice, moesin completely disappeared without affecting the expression levels or subcellular distribution of ezrin and radixin. Also, in platelets, fibroblasts, and mast cells isolated from moesin-deficient mice, targeted disruption of the moesin gene did not affect their ERM-dependent functions, i.e. platelet aggregation, stress fiber/focal contact formation of fibroblasts, and microvillar formation of mast cells, without compensatory up-regulation of ezrin or radixin. These findings favor the notion that ERM proteins are functionally redundant at the cellular as well as the whole body level.
- Tsukita S, Yonemura S
- Cortical actin organization: lessons from ERM (ezrin/radixin/moesin) proteins.
- J Biol Chem. 1999; 274: 34507-10
- Yin HL, Stull JT
- Proteins that regulate dynamic actin remodeling in response to membrane signaling minireview series.
- J Biol Chem. 1999; 274: 32529-30
- Matsui T et al.
- Rho-kinase phosphorylates COOH-terminal threonines of ezrin/radixin/moesin (ERM) proteins and regulates their head-to-tail association.
- J Cell Biol. 1998; 140: 647-57
- Display abstract
The ezrin/radixin/moesin (ERM) proteins are involved in actin filament/plasma membrane interaction that is regulated by Rho. We examined whether ERM proteins are directly phosphorylated by Rho-associated kinase (Rho-kinase), a direct target of Rho. Recombinant full-length and COOH-terminal half radixin were incubated with constitutively active catalytic domain of Rho-kinase, and approximately 30 and approximately 100% of these molecules, respectively, were phosphorylated mainly at the COOH-terminal threonine (T564). Next, to detect Rho-kinase-dependent phosphorylation of ERM proteins in vivo, we raised a mAb that recognized the T564-phosphorylated radixin as well as ezrin and moesin phosphorylated at the corresponding threonine residue (T567 and T558, respectively). Immunoblotting of serum-starved Swiss 3T3 cells with this mAb revealed that after LPA stimulation ERM proteins were rapidly phosphorylated at T567 (ezrin), T564 (radixin), and T558 (moesin) in a Rho-dependent manner and then dephosphorylated within 2 min. Furthermore, the T564 phosphorylation of recombinant COOH-terminal half radixin did not affect its ability to bind to actin filaments in vitro but significantly suppressed its direct interaction with the NH2-terminal half of radixin. These observations indicate that the Rho-kinase-dependent phosphorylation interferes with the intramolecular and/ or intermolecular head-to-tail association of ERM proteins, which is an important mechanism of regulation of their activity as actin filament/plasma membrane cross-linkers.
- Murthy A et al.
- NHE-RF, a regulatory cofactor for Na(+)-H+ exchange, is a common interactor for merlin and ERM (MERM) proteins.
- J Biol Chem. 1998; 273: 1273-6
- Display abstract
We have identified the human homologue of a regulatory cofactor of Na(+)-H+ exchanger (NHE-RF) as a novel interactor for merlin, the neurofibromatosis 2 tumor suppressor protein. NHE-RF mediates protein kinase A regulation of Na(+)-H+ exchanger NHE3 to which it is thought to bind via one of its two PDZ domains. The carboxyl-terminal region of NHE-RF, downstream of the PDZ domains, interacts with the amino-terminal protein 4.1 domain-containing segment of merlin in yeast two-hybrid assays. This interaction also occurs in affinity binding assays with full-length NHE-RF expressed in COS-7 cells. NHE-RF binds to the related ERM proteins, moesin and radixin. We have localized human NHE-RF to actin-rich structures such as membrane ruffles, microvilli, and filopodia in HeLa and COS-7 cells, where it co-localizes with merlin and moesin. These findings suggest that hNHE-RF and its binding partners may participate in a larger complex (one component of which might be a Na(+)-H+ exchanger) that could be crucial for the actin filament assembly activated by the ERM proteins and for the tumor suppressor function of merlin.
- Shaw RJ, Henry M, Solomon F, Jacks T
- RhoA-dependent phosphorylation and relocalization of ERM proteins into apical membrane/actin protrusions in fibroblasts.
- Mol Biol Cell. 1998; 9: 403-19
- Display abstract
The ERM proteins (ezrin, radixin, and moesin) are a group of band 4. 1-related proteins that are proposed to function as membrane/cytoskeletal linkers. Previous biochemical studies have implicated RhoA in regulating the association of ERM proteins with their membrane targets. However, the specific effect and mechanism of action of this regulation is unclear. We show that lysophosphatidic acid stimulation of serum-starved NIH3T3 cells resulted in relocalization of radixin into apical membrane/actin protrusions, which was blocked by inactivation of Rho by C3 transferase. An activated allele of RhoA, but not Rac or CDC42Hs, was sufficient to induce apical membrane/actin protrusions and localize radixin or moesin into these structures in both Rat1 and NIH3T3 cells. Lysophosphatidic acid treatment led to phosphorylation of radixin preceding its redistribution into apical protrusions. Significantly, cotransfection of RhoAV14 or C3 transferase with radixin and moesin revealed that RhoA activity is necessary and sufficient for their phosphorylation. These findings reveal a novel function of RhoA in reorganizing the apical actin cytoskeleton and suggest that this function may be mediated through phosphorylation of ERM proteins.
- Simons PC, Pietromonaco SF, Reczek D, Bretscher A, Elias L
- C-terminal threonine phosphorylation activates ERM proteins to link the cell's cortical lipid bilayer to the cytoskeleton.
- Biochem Biophys Res Commun. 1998; 253: 561-5
- Display abstract
The plasma membrane consists of a lipid bilayer with integral membrane proteins stabilized by regulated linkages to the cortical actin cytoskeleton. The regulation is necessary for cells to change shape ormigrate. The ERM (ezrin-radixin-moesin) proteins are believed to provide such links, with the N-terminal halves associating with integral membrane proteins, either directly or indirectly through adapter molecules like EBP50 (ERM binding phosphoprotein, 50 kDa), and their C-terminal halves associating with F-actin. However, isolated ERM proteins largely exist in a dormant state by virtue of an intramolecular interaction between amino- and carboxyl-terminal domains, thereby masking membrane and cytoskeletal association sites. C-terminal threonine phosphorylation of a fragment of radixin has been found to destroy its ability to bind the amino-terminal domain without affecting the C-terminal F-actin binding site. Here we show that C-terminal phosphorylation of full-length, dormant ezrin and moesin by protein kinase C-theta simultaneously unmasks both the F-actin and EBP50 binding sites. Increased phosphorylation of moesin in cells correlated with increased association of moesin with the cortical actin cytoskeleton. These results show that activation of ERM proteins can be accomplished by phosphorylation of a single C-terminal threonine residue.
- Xu HM, Gutmann DH
- Merlin differentially associates with the microtubule and actin cytoskeleton.
- J Neurosci Res. 1998; 51: 403-15
- Display abstract
The neurofibromatosis 2 (NF2) suppressor gene encodes a protein termed merlin (or schwannomin) with sequence similarity to a family of proteins that link the actin cytoskeleton to cell surface glycoproteins. Members of this ERM family of proteins include ezrin, radixin, and moesin. These proteins contain a carboxyl (C-) terminus actin binding site. In contrast to the ERM proteins, merlin lacks the conventional C-terminal actin binding site, but still localizes to the ruffling edge of plasma membranes. In this study, we investigate the ability of merlin to interact with actin through a nonconventional actin binding domain. We demonstrate for the first time that merlin can associate with polymerized actin in vitro by virtue of an amino (N-) terminal actin binding domain including residues 178-367. Merlin actin binding is not affected by several naturally-occurring NF2 patient mutations or alternatively spliced isoforms. These results suggest that merlin, like other ERM proteins, can directly interact with the actin cytoskeleton. In addition, merlin associates with polymerized microtubules in vitro using a novel microtubule binding region in the N-terminal region of merlin that is masked in the full-length merlin molecule, such that wild-type functional merlin in the "closed" conformation fails to bind polymerized microtubules. These microtubule association results confirm the notion that merlin exists in "open" and "closed" conformations relevant to its function as a negative growth regulator.
- Reczek D, Berryman M, Bretscher A
- Identification of EBP50: A PDZ-containing phosphoprotein that associates with members of the ezrin-radixin-moesin family.
- J Cell Biol. 1997; 139: 169-79
- Display abstract
Members of the ezrin-radixin-moesin (ERM) family of membrane-cytoskeletal linking proteins have NH2- and COOH-terminal domains that associate with the plasma membrane and the actin cytoskeleton, respectively. To search for ERM binding partners potentially involved in membrane association, tissue lysates were subjected to affinity chromatography on the immobilized NH2-terminal domains of ezrin and moesin, which comprise the ezrin-radixin-moesin-association domain (N-ERMAD). A collection of polypeptides at 50-53 kD from human placenta and at 58-59 kD from bovine brain bound directly to both N-ERMADs. The 50-53-kD placental proteins migrated as a major 50-kD species after phosphatase treatment, indicating that the heterogeneity is due to different phosphorylation states. We refer to these polypeptides as ERM-binding phosphoprotein 50 (EBP50). Sequence analysis of human EBP50 was used to identify an approximately 2-kb human cDNA that encodes a 357-residue polypeptide. Recombinant EBP50 binds tightly to the N-ERMADs of ezrin and moesin. Peptide sequences from the brain candidate indicated that it is closely related to EBP50. EBP50 has two PSD-95/DlgA/ZO-1-like (PDZ) domains and is most likely a homologue of rabbit protein cofactor, which is involved in the protein kinase A regulation of the renal brush border Na+/H+ exchanger. EBP50 is widely distributed in tissues, and is particularly enriched in those containing polarized epithelia. Immunofluorescence microscopy of cultured cells and tissues revealed that EBP50 colocalizes with actin and ezrin in the apical microvilli of epithelial cells, and immunoelectron microscopy demonstrated that it is specifically associated with the microvilli of the placental syncytiotrophoblast. Moreover, EBP50 and ezrin can be coimmunoprecipitated as a complex from isolated human placental microvilli. These findings show that EBP50 is a physiologically relevant ezrin binding protein. Since PDZ domains are known to mediate associations with integral membrane proteins, one mode of membrane attachment of ezrin is likely to be mediated through EBP50.
- Tsukita S, Yonemura S, Tsukita S
- ERM proteins: head-to-tail regulation of actin-plasma membrane interaction.
- Trends Biochem Sci. 1997; 22: 53-8
- Display abstract
ERM (ezrin/radixin/moesin) proteins crosslink actin filaments with plasma membranes. The carboxyl termini of these proteins bind actin filaments, while the amino termini bind plasma membranes using a binding partner, such as CD44. Specific signals activate ERM proteins to bind actin filaments and the plasma membrane; these include phosphoinositides and/or phosphorylation mechanisms, which might be located downstream from the Rho-dependent pathway.
- Hirao M et al.
- Regulation mechanism of ERM (ezrin/radixin/moesin) protein/plasma membrane association: possible involvement of phosphatidylinositol turnover and Rho-dependent signaling pathway.
- J Cell Biol. 1996; 135: 37-51
- Display abstract
The ERM proteins, ezrin, radixin, and moesin, are involved in the actin filament/plasma membrane interaction as cross-linkers. CD44 has been identified as one of the major membrane binding partners for ERM proteins. To examine the CD44/ERM protein interaction in vitro, we produced mouse ezrin, radixin, moesin, and the glutathione-S-transferase (GST)/CD44 cytoplasmic domain fusion protein (GST-CD44cyt) by means of recombinant baculovirus infection, and constructed an in vitro assay for the binding between ERM proteins and the cytoplasmic domain of CD44. In this system, ERM proteins bound to GST-CD44cyt with high affinity (Kd of moesin was 9.3 +/- 1.6nM) at a low ionic strength, but with low affinity at a physiological ionic strength. However, in the presence of phosphoinositides (phosphatidylinositol [PI], phosphatidylinositol 4-monophosphate [4-PIP], and phosphatidylinositol 4.5-bisphosphate [4,5-PIP2]), ERM proteins bound with a relatively high affinity to GST-CD44cyt even at a physiological ionic strength: 4,5-PIP2 showed a marked effect (Kd of moesin in the presence of 4,5-PIP2 was 9.3 +/- 4.8 nM). Next, to examine the regulation mechanism of CD44/ERM interaction in vivo, we reexamined the immunoprecipitated CD44/ERM complex from BHK cells and found that it contains Rho-GDP dissociation inhibitor (GDI), a regulator of Rho GTPase. We then evaluated the involvement of Rho in the regulation of the CD44/ERM complex formation. When recombinant ERM proteins were added and incubated with lysates of cultured BHK cells followed by centrifugation, a portion of the recombinant ERM proteins was recovered in the insoluble fraction. This binding was enhanced by GTP gamma S and markedly suppressed by C3 toxin, a specific inhibitor of Rho, indicating that the GTP form of Rho in the lysate is required for this binding. A mAb specific for the cytoplasmic domain of CD44 also markedly suppressed this binding, identifying most of the binding partners for exogenous ERM proteins in the insoluble fraction as CD44. Consistent with this binding analysis, in living BHK cells treated with C3 toxin, most insoluble ERM proteins moved to soluble compartments in the cytoplasm, leaving CD44 free from ERM. These findings indicate that Rho regulates the CD44/ERM complex formation in vivo and that the phosphatidylinositol turnover may be involved in this regulation mechanism.
- Nakamura H, Ozawa H
- Immunolocalization of CD44 and the ERM family in bone cells of mouse tibiae.
- J Bone Miner Res. 1996; 11: 1715-22
- Display abstract
We studied the immunohistochemical localization of CD44, hyaluronate receptor, and the ezrin-radixin-moesin (ERM) family, actin binding proteins, in bone cells using confocal laser scanning microscopy and transmission electron microscopy to clarify the mechanism of the organization of their cytoskeletons. In osteoclasts, intense immunoreactivity to CD44 could be detected on their basolateral plasma membranes. There was less reactivity observed in the area of the plasma membrane in direct contact with the bone surface. The immunogold electron-microscopical method revealed that CD44 was mainly localized on the microvilli of the basolateral plasma membrane. The plasma membrane of the clear zone and the ruffled border were not immunolabeled with CD44. As for the ERM family, the basolateral plasma membrane of osteoclasts was stained with antimoesin monoclonal antibody, but not with ezrin or radixin. In osteoblasts attached to the bone surface, immunoreactivity to CD44 was restricted to their cytoplasmic processes. They showed immunoreactivities to radixin and moesin on the cytoplasmic side of their plasma membrane when in contact with each other. However, although osteocytes in the bone matrix demonstrate an intense immunolabeling with CD44 on their plasma membrane, they scarcely show immunoreactivity to the ERM family. These findings suggest that: (1) the CD44-moesin-actin filament system is involved in the organization of cytoskeletons in the basolateral plasma membrane of osteoclasts; and (2) other mechanisms, rather than the CD44 and the ERM family, may be involved in the cells of osteoblast lineage.
- Tsukita S
- [ERM (ezrin/radixin/moesin) as crosslinkers between actin filaments and plasma membranes]
- Tanpakushitsu Kakusan Koso. 1996; 41: 1899-905
- Pestonjamasp K, Amieva MR, Strassel CP, Nauseef WM, Furthmayr H, Luna EJ
- Moesin, ezrin, and p205 are actin-binding proteins associated with neutrophil plasma membranes.
- Mol Biol Cell. 1995; 6: 247-59
- Display abstract
Actin-binding proteins in bovine neutrophil plasma membranes were identified using blot overlays with 125I-labeled F-actin. Along with surface-biotinylated proteins, membranes were enriched in major actin-binding polypeptides of 78, 81, and 205 kDa. Binding was specific for F-actin because G-actin did not bind. Further, unlabeled F-actin blocked the binding of 125I-labeled F-actin whereas other acidic biopolymers were relatively ineffective. Binding also was specifically inhibited by myosin subfragment 1, but not by CapZ or plasma gelsolin, suggesting that the membrane proteins, like myosin, bind along the sides of the actin filaments. The 78- and 81-kDa polypeptides were identified as moesin and ezrin, respectively, by co-migration on sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoprecipitation with antibodies specific for moesin and ezrin. Although not present in detectable amounts in bovine neutrophils, radixin (a third and closely related member of this gene family) also bound 125I-labeled F-actin on blot overlays. Experiments with full-length and truncated bacterial fusion proteins localized the actin-binding site in moesin to the extreme carboxy terminus, a highly conserved sequence. Immunofluorescence micrographs of permeabilized cells and cell "footprints" showed moesin co-localization with actin at the cytoplasmic surface of the plasma membrane, consistent with a role as a membrane-actin-linking protein.
- Hitt AL, Luna EJ
- Membrane interactions with the actin cytoskeleton.
- Curr Opin Cell Biol. 1994; 6: 120-30
- Display abstract
Recent advances have been made in our understanding of the direct binding of actin to integral membrane proteins. New information has been obtained about indirect actin-membrane associations through spectrin superfamily members and through proteins at the cytoplasmic surfaces of focal contacts and adherens junctions.
- Tsukita S, Oishi K, Sato N, Sagara J, Kawai A, Tsukita S
- ERM family members as molecular linkers between the cell surface glycoprotein CD44 and actin-based cytoskeletons.
- J Cell Biol. 1994; 126: 391-401
- Display abstract
The ERM family members, ezrin, radixin, and moesin, localizing just beneath the plasma membranes, are thought to be involved in the actin filament/plasma membrane association. To identify the integral membrane protein directly associated with ERM family members, we performed immunoprecipitation studies using antimoesin mAb and cultured baby hamster kidney (BHK) cells metabolically labeled with [35S]methionine or surface-labeled with biotin. The results indicated that moesin is directly associated with a 140-kD integral membrane protein. Using BHK cells as antigens, we obtained a mAb that recognized the 140-kD membrane protein. We next cloned a cDNA encoding the 140-kD membrane protein and identified it as CD44, a broadly distributed cell surface glycoprotein. Immunoprecipitation with various anti-CD44 mAbs showed that ezrin and radixin, as well as moesin, are associated with CD44, not only in BHK cells, but also in mouse L fibroblasts. Furthermore, immunofluorescence microscopy revealed that in both BHK and L cells, the Triton X-100-insoluble CD44 is precisely colocalized with ERM family members. We concluded that ERM family members work as molecular linkers between the cytoplasmic domain of CD44 and actin-based cytoskeletons.
- Turunen O, Wahlstrom T, Vaheri A
- Ezrin has a COOH-terminal actin-binding site that is conserved in the ezrin protein family.
- J Cell Biol. 1994; 126: 1445-53
- Display abstract
Ezrin, previously also known as cytovillin, p81, and 80K, is a cytoplasmic protein enriched in microvilli and other cell surface structures. Ezrin is postulated to have a membrane-cytoskeleton linker role. Recent findings have also revealed that the NH2-terminal domain of ezrin is associated with the plasma membrane and the COOH-terminal domain with the cytoskeleton (Algrain, M., O. Turunen, A. Vaheri, D. Louvard, and M. Arpin. 1993. J. Cell Biol. 120: 129-139). Using bacterially expressed fragments of ezrin we now demonstrate that ezrin has an actin-binding capability. We used glutathione-S-transferase fusion proteins of truncated ezrin in affinity chromatography to bind actin from the cell extract or purified rabbit muscle actin. We detected a binding site for filamentous actin that was localized to the COOH-terminal 34 amino acids of ezrin. No binding of monomeric actin was detected in the assay. The region corresponding to the COOH-terminal actin-binding site in ezrin is highly conserved in moesin, actin-capping protein radixin and EM10 protein of E. multilocularis, but not in merlin/schwannomin. Consequently, this site is a potential actin-binding site also in the other members of the protein family. Furthermore, the actin-binding site in ezrin shows sequence homology to the actin-binding site in the COOH terminus of the beta subunit of the actin-capping protein CapZ and one of the potential actin-binding sites in myosin heavy chain. The actin-binding capability of ezrin supports its proposed role as a membrane-cytoskeleton linker.
- Goodlad GA, Clark CM
- Actin and actin-binding proteins in plasma membranes derived from Walker 256 ascites or solid tumour cells.
- Biochim Biophys Acta. 1993; 1145: 177-9
- Display abstract
Plasma membranes from Walker 256 carcinoma cells grown ascitically or as a solid tumour were examined with respect to actin content, [3H]cytochalasin B-binding and the binding of 125I-labelled G-actin to membrane proteins separated by SDS-PAGE. Differences were observed both in cytochalasin B-binding to membrane actin and affinity of 125I-labelled G-actin for specific membrane proteins.
- Furuhashi K, Inagaki M, Hatano S
- [Functional regulation of cytoskeleton by phosphorylation: actin phosphorylation]
- Seikagaku. 1992; 64: 255-9
- Gould KL, Bretscher A, Esch FS, Hunter T
- cDNA cloning and sequencing of the protein-tyrosine kinase substrate, ezrin, reveals homology to band 4.1.
- EMBO J. 1989; 8: 4133-42
- Display abstract
Ezrin is a component of the microvilli of intestinal epithelial cells and serves as a major cytoplasmic substrate for certain protein-tyrosine kinases. We have cloned and sequenced a human ezrin cDNA and report here the entire protein sequence derived from the nucleotide sequence of the cDNA as well as from partial direct protein sequencing. The deduced protein sequence indicates that ezrin is a highly charged protein with an overall pI of 6.1 and a calculated molecular mass of 69,000. The cDNA clone was used to survey the distribution of the ezrin transcript, and the 3.2 kb ezrin mRNA was found to be expressed in the same tissues that are known to express the protein and at the same relative levels. Highest expression was found in intestine, kidney and lung. The cDNA clone hybridized to DNAs from widely divergent organisms indicating that its sequence is highly conserved throughout evolution. The amino acid sequence of ezrin revealed a high degree of similarity within its N-terminal domain to the erythrocyte cytoskeletal protein, band 4.1 and secondary structure predictions indicate that a second region of ezrin contains a long alpha-helix, a feature also common to band 4.1. The structural similarity of ezrin to band 4.1 suggests a mechanism for the observed localization to the membrane, and a role for ezrin in modulating the association of the cortical cytoskeleton with the plasma membrane.
- Keresztes M
- Three actin-binding proteins are developmentally regulated in rat liver plasma membranes.
- FEBS Lett. 1989; 249: 51-5
- Display abstract
Actin-binding membrane proteins (linking microfilaments to the cell membrane) are involved in cytoskeleton-membrane interactions which are supposed to undergo profound changes during cell proliferation and development. In this study 8 polypeptides were shown to bind F-actin directly in the liver cell membranes of mature rats. From these, the abundance of three polypeptides, of 130, 50 and 36 kDa, was observed to increase considerably during postnatal development, which indicates a developmental change in the cytoskeleton-membrane interactions.
- Disease (disease genes where sequence variants are found in this domain)
-
SwissProt sequences and OMIM curated human diseases associated with missense mutations within the B41 domain.
Protein Disease Merlin (P35240) (SMART) OMIM:101000: Neurofibromatosis, type 2 ; Meningioma, NF2-related, sporadic Schwannoma, sporadic ; Neurolemmomatosis ; Malignant mesothelioma, sporadic Unconventional myosin-VIIa (Q13402) (SMART) OMIM:276903: Usher syndrome, type 1B ; Deafness, autosomal recessive 2, neurosensory
OMIM:600060: Deafness, autosomal dominant 11, neurosensory
OMIM:601317: - Metabolism (metabolic pathways involving proteins which contain this domain)
-
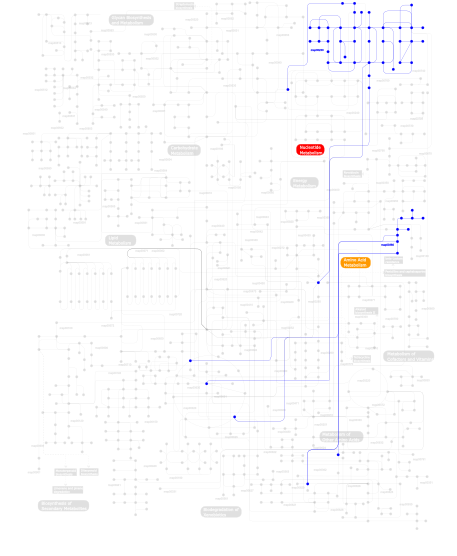
Click the image to view the interactive version of the map in iPath% proteins involved KEGG pathway ID Description 12.61 map04670 Leukocyte transendothelial migration 11.76 map04630 Jak-STAT signaling pathway 11.76 map04920 Adipocytokine signaling pathway 11.34 map04810 Regulation of actin cytoskeleton 10.92 map04510 Focal adhesion 10.50 map04530 Tight junction 3.78 map04012 ErbB signaling pathway 3.78 map05222 Small cell lung cancer 3.78 map04370 VEGF signaling pathway 3.78 map04360 Axon guidance 3.36 map04912 GnRH signaling pathway 3.36 map04020 Calcium signaling pathway 3.36 map04650 Natural killer cell mediated cytotoxicity 2.94  map00380
map00380Tryptophan metabolism 2.52 map04520 Adherens junction 0.42  map00230
map00230Purine metabolism This information is based on mapping of SMART genomic protein database to KEGG orthologous groups. Percentage points are related to the number of proteins with B41 domain which could be assigned to a KEGG orthologous group, and not all proteins containing B41 domain. Please note that proteins can be included in multiple pathways, ie. the numbers above will not always add up to 100%.
- Structure (3D structures containing this domain)
3D Structures of B41 domains in PDB
PDB code Main view Title 1e5w 
Structure of isolated FERM domain and first long helix of moesin 1ef1 
CRYSTAL STRUCTURE OF THE MOESIN FERM DOMAIN/TAIL DOMAIN COMPLEX 1gc6 
CRYSTAL STRUCTURE OF THE RADIXIN FERM DOMAIN COMPLEXED WITH INOSITOL-(1,4,5)-TRIPHOSPHATE 1gc7 
CRYSTAL STRUCTURE OF THE RADIXIN FERM DOMAIN 1gg3 
CRYSTAL STRUCTURE OF THE PROTEIN 4.1R MEMBRANE BINDING DOMAIN 1h4r 
Crystal Structure of the FERM domain of Merlin, the Neurofibromatosis 2 Tumor Suppressor Protein. 1isn 
Crystal structure of merlin FERM domain 1j19 
Crystal structure of the radxin FERM domain complexed with the ICAM-2 cytoplasmic peptide 1mix 
Crystal structure of a FERM domain of Talin 1miz 
Crystal structure of an integrin beta3-talin chimera 1mk7 
CRYSTAL STRUCTURE OF AN INTEGRIN BETA3-TALIN CHIMERA 1mk9 
CRYSTAL STRUCTURE OF AN INTEGRIN BETA3-TALIN CHIMERA 1ni2 
Structure of the active FERM domain of Ezrin 1sgh 
Moesin FERM domain bound to EBP50 C-terminal peptide 1y19 
Structural basis for phosphatidylinositol phosphate kinase type I-gamma binding to talin at focal adhesions 2aeh 
Focal adhesion kinase 1 2al6 
FERM domain of Focal Adhesion Kinase 2d10 
Crystal structure of the Radixin FERM domain complexed with the NHERF-1 C-terminal tail peptide 2d11 
Crystal structure of the Radixin FERM domain complexed with the NHERF-2 C-terminal tail peptide 2d2q 
Crystal structure of the dimerized radixin FERM domain 2ems 
Crystal Structure Analysis of the radixin FERM domain complexed with adhesion molecule CD43 2emt 
Crystal Structure Analysis of the radixin FERM domain complexed with adhesion molecule PSGL-1 2he7 
FERM domain of EPB41L3 (DAL-1) 2hrj 
NMR solution structure of the F2 subdomain of talin 2i1j 
Moesin from Spodoptera frugiperda at 2.1 angstroms resolution 2i1k 
Moesin from Spodoptera frugiperda reveals the coiled-coil domain at 3.0 angstrom resolution 2j0j 
Crystal structure of a fragment of focal adhesion kinase containing the FERM and kinase domains. 2j0k 
Crystal structure of a fragment of focal adhesion kinase containing the FERM and kinase domains. 2j0m 
Crystal structure a two-chain complex between the FERM and kinase domains of focal adhesion kinase. 2rq1 
Solution structure of the 4.1R FERM alpha lobe domain 2yvc 
Crystal structure of the Radixin FERM domain complexed with the NEP cytoplasmic tail 2zpy 
Crystal structure of the mouse radxin FERM domain complexed with the mouse CD44 cytoplasmic peptide 3au4 
Structure of the human myosin-X MyTH4-FERM cassette bound to its specific cargo, DCC 3au5 
Structure of the human myosin-X MyTH4-FERM cassette 3bin 
Structure of the DAL-1 and TSLC1 (372-383) complex 3g9w 
Crystal Structure of Talin2 F2-F3 in Complex with the Integrin Beta1D Cytoplasmic Tail 3ivf 
Crystal structure of the talin head FERM domain 3pvl 
Structure of myosin VIIa MyTH4-FERM-SH3 in complex with the CEN1 of Sans 3pzd 
Structure of the myosin X MyTH4-FERM/DCC complex 3qij 
Primitive-monoclinic crystal structure of the FERM domain of protein 4.1R 3u7d 
Crystal structure of the KRIT1/CCM1 FERM domain in complex with the heart of glass (HEG1) cytoplasmic tail 3u8z 
human merlin FERM domain 3wa0 
3WA0 3x23 
3X23 3zdt 
Crystal structure of basic patch mutant FAK FERM domain FAK31- 405 K216A, K218A, R221A, K222A 4cye 
Crystal structure of avian FAK FERM domain FAK31-405 at 3.2A 4dxa 
Co-crystal structure of Rap1 in complex with KRIT1 4eku 
Crystal Structure of FERM Domain of Proline-rich Tyrosine Kinase 2 4f7g 
Crystal structure of talin autoinhibition complex 4gxb 
Structure of the SNX17 atypical FERM domain bound to the NPxY motif of P-selectin 4hdo 
Crystal structure of the binary Complex of KRIT1 bound to the Rap1 GTPase 4hdq 
Crystal Structure of the Ternary Complex of KRIT1 bound to both the Rap1 GTPase and the Heart of Glass (HEG1) cytoplasmic tail 4ny0 
Crystal structure of FERM domain of human focal adhesion kinase 4p7i 
Crystal structure of the Merlin FERM/DCAF1 complex 4po6 
Crystal structure of the human TYK2 FERM and SH2 domains with an IFNAR1 intracellular peptide 4rm8 
4RM8 4rm9 
4RM9 4rma 
4RMA 4tkn 
4TKN 4yl8 
4YL8 4z32 
4Z32 4zri 
4ZRI 4zrj 
4ZRJ 4zrk 
4ZRK 5d68 
5D68 5ejq 
5EJQ 5ejr 
5EJR 5ejs 
5EJS 5ejy 
5EJY 5f3y 
5F3Y 5ixd 
5IXD 5ixi 
5IXI 5l04 
5L04 - Links (links to other resources describing this domain)
-
PROSITE BAND_41_3 INTERPRO IPR019749 PFAM Band_41

