The domain within your query sequence starts at position 352 and ends at position 393; the E-value for the PAC domain shown below is 1e-12.
SEAHESLMRTSSKREALEKTMRNKMDGEMRRLQDFNRDLRER
The domain was found using the schnipsel database
PACMotif C-terminal to PAS motifs (likely to contribute to PAS structural domain) |
|---|
| SMART accession number: | SM00086 |
|---|---|
| Description: | PAC motif occurs C-terminal to a subset of all known PAS motifs. It is proposed to contribute to the PAS domain fold. |
| Interpro abstract (IPR001610): | PAC motifs occur C-terminal to a subset of all known PAS motifs (see IPR000014 ). It is proposed to contribute to the PAS domain fold [ (PUBMED:9301332) (PUBMED:7756254) (PUBMED:9382818) ]. |
| Family alignment: |
There are 401760 PAC domains in 251051 proteins in SMART's nrdb database.
Click on the following links for more information.
- Evolution (species in which this domain is found)
-
Taxonomic distribution of proteins containing PAC domain.
This tree includes only several representative species. The complete taxonomic breakdown of all proteins with PAC domain is also avaliable.
Click on the protein counts, or double click on taxonomic names to display all proteins containing PAC domain in the selected taxonomic class.
- Literature (relevant references for this domain)
-
Primary literature is listed below; Automatically-derived, secondary literature is also avaliable.
- Pellequer JL, Wager-Smith KA, Kay SA, Getzoff ED
- Photoactive yellow protein: a structural prototype for the three-dimensional fold of the PAS domain superfamily.
- Proc Natl Acad Sci U S A. 1998; 95: 5884-90
- Display abstract
PAS domains are found in diverse proteins throughout all three kingdoms of life, where they apparently function in sensing and signal transduction. Although a wealth of useful sequence and functional information has become recently available, these data have not been integrated into a three-dimensional (3D) framework. The very early evolutionary development and diverse functions of PAS domains have made sequence analysis and modeling of this protein superfamily challenging. Limited sequence similarities between the approximately 50-residue PAS repeats and one region of the bacterial blue-light photosensor photoactive yellow protein (PYP), for which ground-state and light-activated crystallographic structures have been determined to high resolution, originally were identified in sequence searches using consensus sequence probes from PAS-containing proteins. Here, we found that by changing a few residues particular to PYP function, the modified PYP sequence probe also could select PAS protein sequences. By mapping a typical approximately 150-residue PAS domain sequence onto the entire crystallographic structure of PYP, we show that the PAS sequence similarities and differences are consistent with a shared 3D fold (the PAS/PYP module) with obvious potential for a ligand-binding cavity. Thus, PYP appears to prototypically exhibit all the major structural and functional features characteristic of the PAS domain superfamily: the shared PAS/PYP modular domain fold of approximately 125-150 residues, a sensor function often linked to ligand or cofactor (chromophore) binding, and signal transduction capability governed by heterodimeric assembly (to the downstream partner of PYP). This 3D PAS/PYP module provides a structural model to guide experimental testing of hypotheses regarding ligand-binding, dimerization, and signal transduction.
- Rutila JE, Suri V, Le M, So WV, Rosbash M, Hall JC
- CYCLE is a second bHLH-PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless.
- Cell. 1998; 93: 805-14
- Display abstract
We report the identification, characterization, and cloning of another novel Drosophila clock gene, cycle (cyc). Homozygous cyc flies are completely arrhythmic. Heterozygous cyc/+ flies are rhythmic but have altered periods, indicating that the cyc locus has a dosage effect on period. The molecular circadian phenotype of homozygous cyc flies is like homozygous Clk flies presented in the accompanying paper: mutant flies have little or no transcription of the per and tim genes. Cloning of the gene indicates that it also encodes a bHLH-PAS transcription factor and is a Drosophila homolog of the human protein BMAL1. cyc is a nonsense mutation, consistent with its strong loss-of-function phenotype. We propose that the CYC:CLK heterodimer binds to per and tim E boxes and makes a major contribution to the circadian transcription of Drosophila clock genes.
- Ponting CP, Aravind L
- PAS: a multifunctional domain family comes to light.
- Curr Biol. 1997; 7: 6747-6747
- Zelzer E, Wappner P, Shilo BZ
- The PAS domain confers target gene specificity of Drosophila bHLH/PAS proteins.
- Genes Dev. 1997; 11: 2079-89
- Display abstract
Trachealess (Trh) and Single-minded (Sim) are highly similar Drosophila bHLH/PAS transcription factors. They activate nonoverlapping target genes and induce diverse cell fates. A single Drosophila gene encoding a bHLH/PAS protein homologous to the vertebrate ARNT protein was isolated and may serve as a partner for both Trh and Sim. We show that Trh and Sim complexes recognize similar DNA-binding sites in the embryo. To examine the basis for their distinct target gene specificity, the activity of Trh-Sim chimeric proteins was monitored in embryos. Replacement of the Trh PAS domain by the analogous region of Sim was sufficient to convert it into a functional Sim protein. The PAS domain thus mediates all the features conferring specificity and the distinct recognition of target genes. The normal expression pattern of additional proteins essential for the activity of the Trh or Sim complexes can be inferred from the induction pattern of target genes and binding-site reporters, triggered by ubiquitous expression of Trh or Sim. We postulate that the capacity of bHLH/PAS heterodimers to associate, through the PAS domain, with additional distinct proteins that bind target-gene DNA, is essential to confer specificity.
- Zhulin IB, Taylor BL, Dixon R
- PAS domain S-boxes in Archaea, Bacteria and sensors for oxygen and redox.
- Trends Biochem Sci. 1997; 22: 331-3
- Borgstahl GE, Williams DR, Getzoff ED
- 1.4 A structure of photoactive yellow protein, a cytosolic photoreceptor: unusual fold, active site, and chromophore.
- Biochemistry. 1995; 34: 6278-87
- Display abstract
A photosensing protein directs light energy captured by its chromophore into a photocycle. The protein's structure must accommodate the photocycle and promote the resulting chemical or conformational changes that lead to signal transduction. The 1.4 A crystallographic structure of photoactive yellow protein, determined by multiple isomorphous replacement methods, provides the first view at atomic resolution of a protein with a photocycle. The alpha/beta fold, which differs from the original chain tracing, shows striking similarity to distinct parts of the signal transduction proteins profilin and the SH2 domain. In the dark state structure of photoactive yellow protein, the novel 4-hydroxycinnamyl chromophore, covalently attached to Cys69, is buried within the major hydrophobic core of the protein and is tethered at both ends by hydrogen bonds. In the active site, the yellow anionic form of the chromophore is stabilized by hydrogen bonds from the side chains of Tyr42 and buried Glu46 to the phenolic oxygen atom and by electrostatic complementarity with the positively charged guanidinium group of Arg52. Thr50 further interlocks Tyr42, Glu46, and Arg52 through a network of active site hydrogen bonds. Arg52, located in a concavity of the protein surface adjacent to the dominant patch of negative electrostatic potential, shields the chromophore from solvent and is positioned to form a gateway for the phototactic signal. Overall, the high-resolution structure of photoactive yellow protein supports a mechanism whereby electrostatic interactions create an active site poised for photon-induced rearrangements and efficient protein-mediated signal transduction.
- Metabolism (metabolic pathways involving proteins which contain this domain)
-
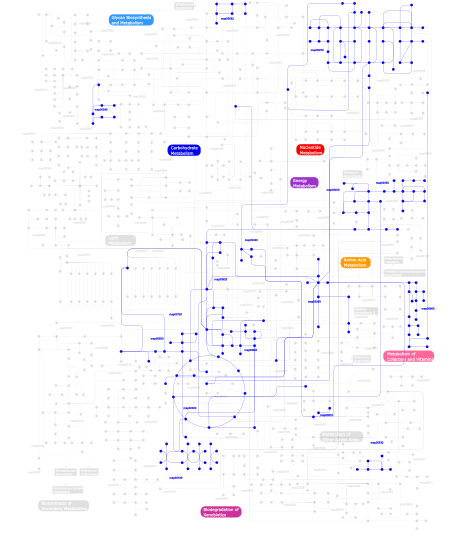
Click the image to view the interactive version of the map in iPath% proteins involved KEGG pathway ID Description 38.51 map02030 Bacterial chemotaxis - General 31.28 map02020 Two-component system - General 9.57 map04710 Circadian rhythm 7.66 map05211 Renal cell carcinoma 2.13 map04150 mTOR signaling pathway 1.70 map03010 Ribosome 1.28  map00230
map00230Purine metabolism 0.64  map00632
map00632Benzoate degradation via CoA ligation 0.64  map00720
map00720Reductive carboxylate cycle (CO2 fixation) 0.64 map04914 Progesterone-mediated oocyte maturation 0.43  map00650
map00650Butanoate metabolism 0.43  map00562
map00562Inositol phosphate metabolism 0.43  map00020
map00020Citrate cycle (TCA cycle) 0.43 map04540 Gap junction 0.43 map04730 Long-term depression 0.21  map00350
map00350Tyrosine metabolism 0.21  map00260
map00260Glycine, serine and threonine metabolism 0.21 map00970 Aminoacyl-tRNA biosynthesis 0.21  map00910
map00910Nitrogen metabolism 0.21  map00620
map00620Pyruvate metabolism 0.21  map00400
map00400Phenylalanine, tyrosine and tryptophan biosynthesis 0.21 map02040 Flagellar assembly 0.21  map00340
map00340Histidine metabolism 0.21 map04810 Regulation of actin cytoskeleton 0.21 map04020 Calcium signaling pathway 0.21 map00633 Trinitrotoluene degradation 0.21  map00190
map00190Oxidative phosphorylation 0.21  map00860
map00860Porphyrin and chlorophyll metabolism 0.21 map04510 Focal adhesion 0.21 map05060 Prion disease 0.21  map00640
map00640Propanoate metabolism 0.21  map00540
map00540Lipopolysaccharide biosynthesis 0.21 map03030 DNA replication This information is based on mapping of SMART genomic protein database to KEGG orthologous groups. Percentage points are related to the number of proteins with PAC domain which could be assigned to a KEGG orthologous group, and not all proteins containing PAC domain. Please note that proteins can be included in multiple pathways, ie. the numbers above will not always add up to 100%.
- Structure (3D structures containing this domain)
3D Structures of PAC domains in PDB
PDB code Main view Title 1byw 
STRUCTURE OF THE N-TERMINAL DOMAIN OF THE HUMAN-ERG POTASSIUM CHANNEL 1dp6 
OXYGEN-BINDING COMPLEX OF FIXL HEME DOMAIN 1dp8 
CRYSTAL STRUCTURE OF THE NITRIC OXIDE BOUND FIXL HEME DOMAIN 1dp9 
CRYSTAL STRUCTURE OF IMIDAZOLE-BOUND FIXL HEME DOMAIN 1drm 
CRYSTAL STRUCTURE OF THE LIGAND FREE BJFIXL HEME DOMAIN 1dwy 
Bovine prion protein fragment 121-230 1dwz 
Bovine prion protein fragment 121-230 1dx0 
BOVINE PRION PROTEIN RESIDUES 23-230 1dx1 
BOVINE PRION PROTEIN RESIDUES 23-230 1g28 
STRUCTURE OF A FLAVIN-BINDING DOMAIN, LOV2, FROM THE CHIMERIC PHYTOCHROME/PHOTOTROPIN PHOTORECEPTOR PHY3 1jnu 
Photoexcited structure of the plant photoreceptor domain, phy3 LOV2 1lsv 
Crystal structure of the CO-bound BjFixL heme domain 1lsw 
Crystal structure of the ferrous BjFixL heme domain 1lsx 
Crystal structure of the methylimidazole-bound BjFixL heme domain 1lt0 
Crystal structure of the CN-bound BjFixL heme domain 1n9l 
Crystal structure of the Phot-LOV1 domain from Chlamydomonas reinhardtii in the dark state. 1n9n 
Crystal structure of the Phot-LOV1 domain from Chlamydomonas reinhardtii in illuminated state. Data set of a single crystal. 1n9o 
Crystal structure of the Phot-LOV1 domain from Chlamydomonas reinhardtii in illuminated state. Composite data set. 1p97 
NMR structure of the C-terminal PAS domain of HIF2a 1s66 
Crystal structure of heme domain of direct oxygen sensor from E. coli 1s67 
Crystal structure of heme domain of direct oxygen sensor from E. coli 1v9y 
Crystal Structure of the heme PAS sensor domain of Ec DOS (ferric form) 1v9z 
Crystal Structure of the heme PAS sensor domain of Ec DOS (Ferrous Form) 1vb6 
Crystal Structure of the heme PAS sensor domain of Ec DOS (oxygen-bound form) 1wa9 
Crystal Structure of the PAS repeat region of the Drosophila clock protein PERIOD 1x0o 
human ARNT C-terminal PAS domain 1xj2 
CO-bound structure of bjFixLH 1xj3 
bjFixLH in unliganded ferrous form 1xj4 
CO-bound structure of BjFixLH 1xj6 
Structure of bjFixLH in the unliganded ferrous form 1y28 
Crystal structure of the R220A metBJFIXL HEME domain 2a24 
HADDOCK Structure of HIF-2a/ARNT PAS-B Heterodimer 2b02 
Crystal Structure of ARNT PAS-B Domain 2cmn 
A Proximal Arginine Residue in the Switching Mechanism of the FixL Oxygen Sensor 2gj3 
Crystal structure of the FAD-containing PAS domain of the protein NifL from Azotobacter vinelandii. 2hv1 
HADDOCK structure of ARNT PAS-B Homodimer 2k7s 
Human ARNT C-Terminal PAS Domain, 3 Residue IB slip 2kdk 
Structure of human circadian clock protein BMAL2 C-terminal PAS domain 2l0w 
Solution NMR structure of the N-terminal PAS domain of HERG potassium channel 2l1m 
Solution structure of the eag domain of the hERG (Kv11.1) K+ channel 2l4r 
NMR solution structure of the N-terminal PAS domain of hERG 2mwg 
2MWG 2owh 
Structure of an early-microsecond photolyzed state of CO-bjFixLH 2owj 
Structure of an early-microsecond photolyzed state of CO-bjFixLH, dark state 2pd7 
2.0 Angstrom Crystal Structure of the Fungal Blue-Light Photoreceptor Vivid 2pd8 
1.8 Angstrom Crystal Structure of the Cys71Ser mutant of Vivid 2pdr 
1.7 Angstrom Crystal Structure of the Photo-excited Blue-light Photoreceptor Vivid 2pdt 
2.3 Angstrom Structure of Phosphodiesterase treated Vivid 2pr5 
Structural Basis for Light-dependent Signaling in the Dimeric LOV Photosensor YtvA (Dark Structure) 2pr6 
Structural Basis for Light-dependent Signaling in the Dimeric LOV Photosensor YtvA (Light Structure) 2v0u 
n- and c-terminal helices of oat lov2 (404-546) are involved in light-induced signal transduction (cryo dark structure of lov2 (404-546)) 2v0w 
N- and C-terminal helices of oat LOV2 (404-546) are involved in light- induced signal transduction (cryo-trapped light structure of LOV2 ( 404-546)) 2v1a 
N- and C-terminal helices of oat LOV2 (404-546) are involved in light-induced signal transduction (room temperature (293K) dark structure of LOV2 (404-546)) 2v1b 
N- and C-terminal helices of oat LOV2 (404-546) are involved in light-induced signal transduction (room temperature (293K) light structure of LOV2 (404-546)) 2vlg 
KinA PAS-A domain, homodimer 2vv6 
BJFIXLH IN FERRIC FORM 2vv7 
BJFIXLH IN UNLIGANDED FERROUS FORM 2vv8 
Molecular mechanism of signal transduction in bjFixL 2wkp 
Structure of a photoactivatable Rac1 containing Lov2 Wildtype 2wkq 
Structure of a photoactivatable Rac1 containing the Lov2 C450A Mutant 2wkr 
Structure of a photoactivatable Rac1 containing the Lov2 C450M Mutant 2z6c 
Crystal structure of LOV1 domain of phototropin1 from Arabidopsis thaliana 2z6d 
Crystal structure of LOV1 domain of phototropin2 from Arabidopsis thaliana 3d72 
1.65 Angstrom crystal structure of the Cys71Val variant in the fungal photoreceptor VVD 3ewk 
Structure of the redox sensor domain of Methylococcus capsulatus (Bath) MmoS 3f1n 
Crystal structure of a high affinity heterodimer of HIF2 alpha and ARNT C-terminal PAS domains, with internally bound ethylene glycol. 3f1o 
Crystal structure of the high affinity heterodimer of HIF2 alpha and ARNT C-terminal PAS domains, with an internally-bound artificial ligand 3f1p 
Crystal structure of a high affinity heterodimer of HIF2 alpha and ARNT C-terminal PAS domains 3gdi 
Mammalian Clock Protein mPER2 - Crystal Struture of a PAS Domain Fragment 3gec 
Crystal structure of a tandem PAS domain fragment of Drosophila PERIOD 3h7w 
Crystal structure of the high affinity heterodimer of HIF2 alpha and ARNT C-terminal PAS domains with the artificial ligand THS017 3h82 
Crystal structure of the high affinity heterodimer of HIF2 alpha and ARNT C-terminal PAS domains with the artificial ligand THS020 3h9w 
Crystal Structure of the N-terminal domain of Diguanylate cyclase with PAS/PAC sensor (Maqu_2914) from Marinobacter aquaeolei, Northeast Structural Genomics Consortium Target MqR66C 3hji 
1.8 Angstrom Crystal Structure of the I74V:I85V Variant of Vivid (VVD). 3hjk 
2.0 Angstrom Structure of the Ile74Val Variant of Vivid (VVD). 3is2 
2.3 Angstrom Crystal Structure of a Cys71 Sulfenic Acid form of Vivid 3k3c 
The N-terminal PAS domain crystal structure of Rv1364c from Mycobacterium tuberculosis at 1.62 3k3d 
The N-terminal PAS domain crystal structure of RV1364C from Mycobacterium Tuberculosis at 2.3 angstrom 3kx0 
Crystal Structure of the PAS domain of Rv1364c 3lyx 
Crystal structure of the PAS domain of the protein CPS_1291 from Colwellia psychrerythraea. Northeast Structural Genomics Consortium target id CsR222B 3mr0 
Crystal Structure of Sensory Box Histidine Kinase/Response Regulator from Burkholderia thailandensis E264 3nja 
The crystal structure of the PAS domain of a GGDEF family protein from Chromobacterium violaceum ATCC 12472. 3p7n 
Crystal structure of light activated transcription factor El222 from Erythrobacter litoralis 3rh8 
Crystal Structure of the Light-state Dimer of Fungal Blue-Light Photoreceptor Vivid 3rty 
Structure of an Enclosed Dimer Formed by The Drosophila Period Protein 3sw1 
Structure of a full-length bacterial LOV protein 3t50 
X-ray structure of the LOV domain from the LOV-HK sensory protein from Brucella abortus (dark state). 3ue6 
The dark structure of the blue-light photoreceptor Aureochrome1 LOV 3ulf 
The light state structure of the blue-light photoreceptor Aureochrome1 LOV 4dj2 
Unwinding the Differences of the Mammalian PERIOD Clock Proteins from Crystal Structure to Cellular Function 4dj3 
Unwinding the Differences of the Mammalian PERIOD Clock Proteins from Crystal Structure to Cellular Function 4eep 
Crystal structure of LOV2 domain of Arabidopsis thaliana phototropin 2 4eer 
Crystal structure of LOV2 domain of Arabidopsis thaliana phototropin 2 C426A mutant 4ees 
Crystal structure of iLOV 4eet 
Crystal structure of iLOV 4eeu 
Crystal structure of phiLOV2.1 4eq1 
Crystal Structure of the ARNT PAS-B homodimer 4f3l 
Crystal Structure of the Heterodimeric CLOCK:BMAL1 Transcriptional Activator Complex 4gcz 
Structure of a blue-light photoreceptor 4ghi 
Crystal structure of the high affinity heterodimer of HIF2 alpha and ARNT C-terminal PAS domains in complex with a benzoxadiazole antagonist 4gs9 
Crystal structure of the high affinity heterodimer of HIF2 alpha and ARNT C-terminal PAS domains in complex with an inactive benzoxadiazole antagonist 4gw9 
Structure of a bacteriophytochrome and light-stimulated protomer swapping with a gene repressor 4h6j 
Identification of Cys 255 in HIF-1 as a novel site for development of covalent inhibitors of HIF-1 /ARNT PasB domain protein-protein interaction. 4hhd 
2.75 Angstrom resolution crystal structure of the A. thaliana LOV2 domain with an extended N-terminal A' helix (cryo dark structure) 4hoi 
Crystal structure of PAS domain from the mouse EAG1 potassium channel 4hp4 
Crystal structure of PAS domain from the fruit-fly ELK potassium channel 4hp9 
Crystal structure of the N-terminal truncated PAS domain from the hERG potassium channel 4hqa 
Crystal structure of PAS domain from the human ERG (hERG) potassium channel 4i5s 
Structure and function of sensor histidine kinase 4kuk 
4KUK 4kuo 
4KUO 4llo 
Structure of the eag domain-CNBHD complex of the mouse EAG1 channel 4lpz 
4LPZ 4nxb 
4NXB 4nxe 
4NXE 4nxf 
4NXF 4nxg 
4NXG 4pky 
4PKY 4r38 
4R38 4r3a 
4R3A 4wf0 
4WF0 4wn5 
4WN5 4xt2 
4XT2 4yx2 
4YX2 4zp4 
4ZP4 4zph 
4ZPH 4zpk 
4ZPK 4zpr 
4ZPR 4zqd 
4ZQD 5a8b 
5A8B 5djt 
5DJT 5dju 
5DJU 5dkk 
5DKK 5dkl 
5DKL 5efw 
5EFW 5hwt 
5HWT 5hwv 
5HWV 5hww 
5HWW 5j3w 
5J3W 5j4e 
5J4E 5j7e 
5J7E 5k7l 
5K7L 5kiz 
5KIZ 5sy5 
5SY5 5sy7 
5SY7 5t0t 
5T0T 5tbm 
5TBM - Links (links to other resources describing this domain)
-
INTERPRO IPR001610

