The domain within your query sequence starts at position 3 and ends at position 215; the E-value for the Glyco_18 domain shown below is 2.24e-47.
NNEITTIEWNDVTLYKAFNDLKNRNSKLKTLLAIGGWNFGTAPFTTMVSTSQNRQTFITS VIKFLRQYGFDGLDLDWEYPGSRGSPPQDKHLFTVLVKEMREAFEQEAIESNRPRLMVTA AVAGGISNIQAGYEIPELSKYLDFIHVMTYDLHGSWEGYTGENSPLYKYPTETGSNAYLN VDYVMNYWKNNGAPAEKLIVGFPEYGHTFILRN
Glyco_18 |
|---|
| SMART accession number: | SM00636 |
|---|---|
| Description: | - |
| Interpro abstract (IPR011583): | O-Glycosyl hydrolases ( EC 3.2.1. ) are a widespread group of enzymes that hydrolyse the glycosidic bond between two or more carbohydrates, or between a carbohydrate and a non-carbohydrate moiety. A classification system for glycosyl hydrolases, based on sequence similarity, has led to the definition of 85 different families [ (PUBMED:7624375) (PUBMED:8535779) ]. This classification is available on the CAZy (CArbohydrate-Active EnZymes) website. Members of this family belong to the chitinase class II group which includes chitinase, chitodextrinase and the killer toxin of Kluyveromyces lactis (Yeast) (Candida sphaerica) and all belong to glycoside hydrolase, family 18 . The chitinases hydrolyse chitin oligosaccharides. However, glycoside hydrolase family 18 also includes chitinase-like proteins, which bind but do not cleave chitin [ (PUBMED:22550243) ]. |
| GO function: | chitin binding (GO:0008061) |
| Family alignment: |
There are 27162 Glyco_18 domains in 25962 proteins in SMART's nrdb database.
Click on the following links for more information.
- Evolution (species in which this domain is found)
-
Taxonomic distribution of proteins containing Glyco_18 domain.
This tree includes only several representative species. The complete taxonomic breakdown of all proteins with Glyco_18 domain is also avaliable.
Click on the protein counts, or double click on taxonomic names to display all proteins containing Glyco_18 domain in the selected taxonomic class.
- Metabolism (metabolic pathways involving proteins which contain this domain)
-
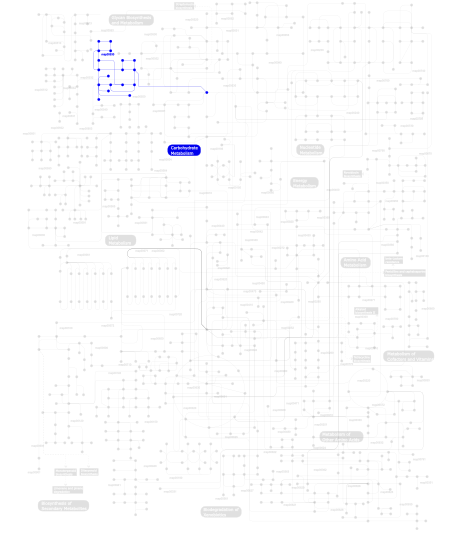
Click the image to view the interactive version of the map in iPath% proteins involved KEGG pathway ID Description 100.00  map00530
map00530Aminosugars metabolism This information is based on mapping of SMART genomic protein database to KEGG orthologous groups. Percentage points are related to the number of proteins with Glyco_18 domain which could be assigned to a KEGG orthologous group, and not all proteins containing Glyco_18 domain. Please note that proteins can be included in multiple pathways, ie. the numbers above will not always add up to 100%.
- Structure (3D structures containing this domain)
3D Structures of Glyco_18 domains in PDB
PDB code Main view Title 1c3f 
Endo-Beta-N-Acetylglucosaminidase H, D130N Mutant 1c8x 
Endo-Beta-N-Acetylglucosaminidase H, D130E Mutant 1c8y 
Endo-Beta-N-Acetylglucosaminidase H, D130A Mutant 1c90 
Endo-Beta-N-Acetylglucosaminidase H, E132Q Mutant 1c91 
Endo-Beta-N-Acetylglucosaminidase H, E132D 1c92 
Endo-Beta-N-Acetylglucosaminidase H, E132A Mutant 1c93 
Endo-Beta-N-Acetylglucosaminidase H, D130N/E132Q Double Mutant 1ctn 
CRYSTAL STRUCTURE OF A BACTERIAL CHITINASE AT 2.3 ANGSTROMS RESOLUTION 1d2k 
C. IMMITIS CHITINASE 1 AT 2.2 ANGSTROMS RESOLUTION 1e15 
Chitinase B from Serratia Marcescens 1e6n 
Chitinase B from Serratia marcescens inactive mutant E144Q in complex with N-acetylglucosamine-pentamer 1e6p 
Chitinase B from Serratia marcescens inactive mutant E144Q 1e6r 
Chitinase B from Serratia marcescens wildtype in complex with inhibitor allosamidin 1e6z 
Chitinase B from Serratia marcescens wildtype in complex with catalytic intermediate 1e9l 
The crystal structure of novel mammalian lectin Ym1 suggests a saccharide binding site 1edq 
CRYSTAL STRUCTURE OF CHITINASE A FROM S. MARCESCENS AT 1.55 ANGSTROMS 1edt 
CRYSTAL STRUCTURE OF ENDO-BETA-N-ACETYLGLUCOSAMINIDASE H AT 1.9 ANGSTROMS RESOLUTION: ACTIVE SITE GEOMETRY AND SUBSTRATE RECOGNITION 1ehn 
CRYSTAL STRUCTURE OF CHITINASE A MUTANT E315Q COMPLEXED WITH OCTA-N-ACETYLCHITOOCTAOSE (NAG)8. 1eib 
CRYSTAL STRUCTURE OF CHITINASE A MUTANT D313A COMPLEXED WITH OCTA-N-ACETYLCHITOOCTAOSE (NAG)8. 1ffq 
CRYSTAL STRUCTURE OF CHITINASE A COMPLEXED WITH ALLOSAMIDIN 1ffr 
CRYSTAL STRUCTURE OF CHITINASE A MUTANT Y390F COMPLEXED WITH HEXA-N-ACETYLCHITOHEXAOSE (NAG)6 1goi 
Crystal structure of the D140N mutant of chitinase B from Serratia marcescens at 1.45 A resolution 1gpf 
CHITINASE B FROM SERRATIA MARCESCENS IN COMPLEX WITH INHIBITOR PSAMMAPLIN 1guv 
Structure of human chitotriosidase 1h0g 
Complex of a chitinase with the natural product cyclopentapeptide argadin from Clonostachys 1h0i 
Complex of a chitinase with the natural product cyclopentapeptide argifin from Gliocladium 1hjv 
Crystal structure of hcgp-39 in complex with chitin tetramer 1hjw 
Crystal structure of hcgp-39 in complex with chitin octamer 1hjx 
Ligand-induced signalling and conformational change of the 39 kD glycoprotein from human articular chondrocytes 1hki 
Crystal structure of human chitinase in complex with glucoallosamidin B 1hkj 
Crystal structure of human chitinase in complex with methylallosamidin 1hkk 
High resoultion crystal structure of human chitinase in complex with allosamidin 1hkm 
High resolution crystal structure of human chitinase in complex with demethylallosamidin 1hvq 
CRYSTAL STRUCTURES OF HEVAMINE, A PLANT DEFENCE PROTEIN WITH CHITINASE AND LYSOZYME ACTIVITY, AND ITS COMPLEX WITH AN INHIBITOR 1itx 
Catalytic Domain of Chitinase A1 from Bacillus circulans WL-12 1jnd 
Crystal structure of imaginal disc growth factor-2 1jne 
Crystal structure of imaginal disc growth factor-2 1k9t 
Chitinase a complexed with tetra-N-acetylchitotriose 1kfw 
Structure of catalytic domain of psychrophilic chitinase B from Arthrobacter TAD20 1kr0 
Hevamine Mutant D125A/Y183F in Complex with Tetra-NAG 1lg1 
CRYSTAL STRUCTURE OF HUMAN CHITOTRIOSIDASE IN COMPLEX WITH CHITOBIOSE 1lg2 
CRYSTAL STRUCTURE OF HUMAN CHITOTRIOSIDASE IN COMPLEX WITH ETHYLENE GLYCOL 1ljy 
Crystal Structure of a Novel Regulatory 40 kDa Mammary Gland Protein (MGP-40) secreted during Involution 1ll4 
STRUCTURE OF C. IMMITIS CHITINASE 1 COMPLEXED WITH ALLOSAMIDIN 1ll6 
STRUCTURE OF THE D169N MUTANT OF C. IMMITIS CHITINASE 1 1ll7 
STRUCTURE OF THE E171Q MUTANT OF C. IMMITIS CHITINASE 1 1llo 
HEVAMINE A (A PLANT ENDOCHITINASE/LYSOZYME) COMPLEXED WITH ALLOSAMIDIN 1lq0 
CRYSTAL STRUCTURE OF HUMAN CHITOTRIOSIDASE AT 2.2 ANGSTROM RESOLUTION 1nar 
CRYSTAL STRUCTURE OF NARBONIN REFINED AT 1.8 ANGSTROMS RESOLUTION 1nh6 
Structure of S. marcescens chitinase A, E315L, complex with hexasaccharide 1nwr 
Crystal structure of human cartilage gp39 (HC-gp39) 1nws 
Crystal structure of human cartilage gp39 (HC-gp39) in complex with chitobiose 1nwt 
Crystal structure of human cartilage gp39 (HC-gp39) in complex with chitopentaose 1nwu 
Crystal structure of human cartilage gp39 (HC-gp39) in complex with chitotetraose 1o6i 
Chitinase B from Serratia marcescens complexed with the catalytic intermediate mimic cyclic dipeptide CI4. 1ogb 
chitinase b from serratia marcescens mutant d142n 1ogg 
chitinase b from serratia marcescens mutant d142n in complex with inhibitor allosamidin 1owq 
Crystal structure of a 40 kDa signalling protein (SPC-40) secreted during involution 1rd6 
Crystal Structure of S. Marcescens Chitinase A Mutant W167A 1sr0 
Crystal structure of signalling protein from sheep(SPS-40) at 3.0A resolution using crystal grown in the presence of polysaccharides 1syt 
Crystal structure of signalling protein from goat SPG-40 in the presense of N,N',N''-triacetyl-chitotriose at 2.6A resolution 1tfv 
CRYSTAL STRUCTURE OF A BUFFALO SIGNALING GLYCOPROTEIN (SPB-40) SECRETED DURING INVOLUTION 1ur8 
Interactions of a family 18 chitinase with the designed inhibitor HM508, and its degradation product, chitobiono-delta-lactone 1ur9 
Interactions of a family 18 chitinase with the designed inhibitor HM508, and its degradation product, chitobiono-delta-lactone 1vf8 
The Crystal Structure of Ym1 at 1.31 A Resolution 1w1p 
Crystal structure of S. marcescens chitinase B in complex with the cyclic dipeptide inhibitor cyclo-(Gly-L-Pro) at 2.1 A resolution 1w1t 
Crystal structure of S. marcescens chitinase B in complex with the cyclic dipeptide inhibitor cyclo-(His-L-Pro) at 1.9 A resolution 1w1v 
Crystal structure of S. marcescens chitinase B in complex with the cyclic dipeptide inhibitor cyclo-(L-Arg-L-Pro) at 1.85 A resolution 1w1y 
Crystal structure of S. marcescens chitinase B in complex with the cyclic dipeptide inhibitor cyclo-(L-Tyr-L-Pro) at 1.85 A resolution 1w9p 
Specificity and affinity of natural product cyclopentapeptide inhibitors against Aspergillus fumigatus, human and bacterial chitinaseFra 1w9u 
Specificity and affnity of natural product cyclopentapeptide inhibitor Argadin against Aspergillus fumigatus chitinase 1w9v 
Specificity and affinity of natural product cyclopentapeptide argifin against Aspergillus fumigatus 1waw 
Specificity and affnity of natural product cyclopentapeptide inhibitor Argadin against human chitinase 1wb0 
specificity and affinity of natural product cyclopentapeptide inhibitor Argifin against human chitinaes 1wno 
Crystal structure of a native chitinase from Aspergillus fumigatus YJ-407 1x6l 
Crystal structure of S. marcescens chitinase A mutant W167A 1x6n 
Crystal structure of S. marcescens chitinase A mutant W167A in complex with allosamidin 1xhg 
Crystal structure of a 40 kDa signalling protein from Porcine (SPP-40) at 2.89A resolution 1xrv 
Crystal Structure of the novel secretory signalling protein from Porcine (SPP-40) at 2.1A resolution. 1zb5 
Recognition of peptide ligands by signalling protein from porcine mammary gland (SPP-40): Crystal structure of the complex of SPP-40 with a peptide Trp-Pro-Trp at 2.45A resolution 1zbc 
Crystal Structure of the porcine signalling protein liganded with the peptide Trp-Pro-Trp (WPW) at 2.3 A resolution 1zbk 
Recognition of specific peptide sequences by signalling protein from sheep mammary gland (SPS-40): Crystal structure of the complex of SPS-40 with a peptide Trp-Pro-Trp at 2.9A resolution 1zbv 
Crystal Structure of the goat signalling protein (SPG-40) complexed with a designed peptide Trp-Pro-Trp at 3.2A resolution 1zbw 
Crystal structure of the complex formed between signalling protein from goat mammary gland (SPG-40) and a tripeptide Trp-Pro-Trp at 2.8A resolution 1zl1 
Crystal structure of the complex of signalling protein from sheep (SPS-40) with a designed peptide Trp-His-Trp reveals significance of Asn79 and Trp191 in the complex formation 1zu8 
Crystal structure of the goat signalling protein with a bound trisaccharide reveals that Trp78 reduces the carbohydrate binding site to half 2a3a 
Crystal structure of Aspergillus fumigatus chitinase B1 in complex with theophylline 2a3b 
Crystal structure of Aspergillus fumigatus chitinase B1 in complex with caffeine 2a3c 
Crystal structure of Aspergillus fumigatus chitinase B1 in complex with pentoxifylline 2a3e 
Crystal structure of Aspergillus fumigatus chitinase B1 in complex with allosamidin 2aos 
Protein-protein Interactions of protective signalling factor: Crystal structure of ternary complex involving signalling protein from goat (SPG-40), tetrasaccharide and a tripeptide Trp-pro-Trp at 2.9 A resolution 2b31 
Crystal structure of the complex formed between goat signalling protein with pentasaccharide at 3.1 A resolution reveals large scale conformational changes in the residues of TIM barrel 2dpe 
Crystal structure of a secretory 40KDA glycoprotein from sheep at 2.0A resolution 2dsu 
Binding of chitin-like polysaccharide to protective signalling factor: Crystal structure of the complex formed between signalling protein from sheep (SPS-40) with a tetrasaccharide at 2.2 A resolution 2dsv 
Interactions of protective signalling factor with chitin-like polysaccharide: Crystal structure of the complex between signalling protein from sheep (SPS-40) and a hexasaccharide at 2.5A resolution 2dsw 
Binding of chitin-like polysaccharides to protective signalling factor: crystal structure of the complex of signalling protein from sheep (SPS-40) with a pentasaccharide at 2.8 A resolution 2dsz 
Three dimensional structure of a goat signalling protein secreted during involution 2dt0 
Crystal structure of the complex of goat signalling protein with the trimer of N-acetylglucosamine at 2.45A resolution 2dt1 
Crystal Structure Of The Complex Of Goat Signalling Protein With Tetrasaccharide At 2.09 A Resolution 2dt2 
Crystal structure of the complex formed between goat signalling protein with pentasaccharide at 2.9A resolution 2dt3 
Crystal structure of the complex formed between goat signalling protein and the hexasaccharide at 2.28 A resolution 2ebn 
CRYSTAL STRUCTURE OF ENDO-BETA-N-ACETYLGLUCOSAMINIDASE F1, AN ALPHA(SLASH)BETA-BARREL ENZYME ADAPTED FOR A COMPLEX SUBSTRATE 2esc 
Crystal structure of a 40 KDa protective signalling protein from Bovine (SPC-40) at 2.1 A resolution 2fdm 
Crystal structure of the ternary complex of signalling glycoprotein frm sheep (SPS-40)with hexasaccharide (NAG6) and peptide Trp-Pro-Trp at 3.0A resolution 2g41 
Crystal structure of the complex of sheep signalling glycoprotein with chitin trimer at 3.0A resolution 2g8z 
Crystal structure of the ternary complex of signalling protein from sheep (SPS-40) with trimer and designed peptide at 2.5A resolution 2gsj 
cDNA cloning and 1.75A crystal structure determination of PPL2, a novel chimerolectin from Parkia platycephala seeds exhibiting endochitinolytic activity 2hvm 
HEVAMINE A AT 1.8 ANGSTROM RESOLUTION 2iuz 
Crystal structure of Aspergillus fumigatus chitinase B1 in complex with C2-dicaffeine 2o92 
Crystal structure of a signalling protein (SPG-40) complex with tetrasaccharide at 3.0A resolution 2o9o 
Crystal Structure of the buffalo Secretory Signalling Glycoprotein at 2.8 A resolution 2olh 
Crystal structure of a signalling protein (SPG-40) complex with cellobiose at 2.78 A resolution 2pi6 
Crystal structure of the sheep signalling glycoprotein (SPS-40) complex with 2-methyl-2-4-pentanediol at 1.65A resolution reveals specific binding characteristics of SPS-40 2qf8 
Crystal structure of the complex of Buffalo Secretory Glycoprotein with tetrasaccharide at 2.8A resolution 2uy2 
ScCTS1_apo crystal structure 2uy3 
ScCTS1_8-chlorotheophylline crystal structure 2uy4 
ScCTS1_acetazolamide crystal structure 2uy5 
ScCTS1_kinetin crystal structure 2wk2 
Chitinase A from Serratia marcescens ATCC990 in complex with Chitotrio-thiazoline dithioamide. 2wly 
Chitinase A from Serratia marcescens ATCC990 in complex with Chitotrio-thiazoline. 2wlz 
Chitinase A from Serratia marcescens ATCC990 in complex with Chitobio- thiazoline. 2wm0 
Chitinase A from Serratia marcescens ATCC990 in complex with Chitobio- thiazoline thioamide. 2xtk 
ChiA1 from Aspergillus fumigatus in complex with acetazolamide 2xuc 
Natural product-guided discovery of a fungal chitinase inhibitor 2xvn 
A. fumigatus chitinase A1 phenyl-methylguanylurea complex 2xvp 
ChiA1 from Aspergillus fumigatus, apostructure 2y8v 
Structure of chitinase, ChiC, from Aspergillus fumigatus. 2ybt 
Crystal structure of human acidic chitinase in complex with bisdionin C 2ybu 
Crystal structure of human acidic chitinase in complex with bisdionin F 3alf 
Crystal Structure of Class V Chitinase from Nicotiana tobaccum 3alg 
Crystal Structure of Class V Chitinase (E115Q mutant) from Nicotiana tobaccum in complex with NAG4 3aqu 
Crystal structure of a class V chitinase from Arabidopsis thaliana 3aro 
Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - apo structure 3arp 
Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - complex structure with DEQUALINIUM 3arq 
Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - complex structure with IDARUBICIN 3arr 
Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - complex structure with PENTOXIFYLLINE 3ars 
Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - apo structure of mutant W275G 3art 
Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - W275G mutant complex structure with DEQUALINIUM 3aru 
Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - W275G mutant complex structure with PENTOXIFYLLINE 3arv 
Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - complex structure with Sanguinarine 3arw 
Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - complex structure with chelerythrine 3arx 
Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - complex structure with Propentofylline 3ary 
Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - complex structure with 2-(imidazolin-2-yl)-5-isothiocyanatobenzofuran 3arz 
Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - complex structure with 2-(imidazolin-2-yl)-5-isothiocyanatobenzofuran 3as0 
Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - W275G mutant complex structure with Sanguinarine 3as1 
Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - W275G mutant complex structure with chelerythrine 3as2 
Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - W275G mutant complex structure with Propentofylline 3as3 
Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - W275G mutant complex structure with 2-(imidazolin-2-yl)-5-isothiocyanatobenzofuran 3b8s 
Crystal structure of wild-type chitinase A from Vibrio harveyi 3b9a 
Crystal structure of Vibrio harveyi chitinase A complexed with hexasaccharide 3b9d 
Crystal structure of Vibrio harveyi chitinase A complexed with pentasaccharide 3b9e 
Crystal structure of inactive mutant E315M chitinase A from Vibrio harveyi 3bxw 
Crystal Structure of Stabilin-1 Interacting Chitinase-Like Protein, SI-CLP 3ch9 
Crystal structure of Aspergillus fumigatus chitinase B1 in complex with dimethylguanylurea 3chc 
Crystal structure of Aspergillus fumigatus chitinase B1 in complex with monopeptide 3chd 
Crystal structure of Aspergillus fumigatus chitinase B1 in complex with dipeptide 3che 
Crystal structure of Aspergillus fumigatus chitinase B1 in complex with tripeptide 3chf 
Crystal structure of Aspergillus fumigatus chitinase B1 in complex with tetrapeptide 3co4 
Crystal structure of a chitinase from Bacteroides thetaiotaomicron 3cz8 
Crystal structure of putative sporulation-specific glycosylase ydhD from Bacillus subtilis 3d5h 
Crystal structure of haementhin from Haemanthus multiflorus at 2.0A resolution: Formation of a novel loop on a TIM barrel fold and its functional significance 3ebv 
Crystal structure of putative Chitinase A from Streptomyces coelicolor. 3fnd 
Crystal structure of a chitinase from Bacteroides thetaiotaomicron 3fxy 
Acidic Mammalian Chinase, Catalytic Domain 3fy1 
The Acidic Mammalian Chitinase catalytic domain in complex with methylallosamidin 3g6l 
The crystal structure of a chitinase CrChi1 from the nematophagous fungus Clonostachys rosea 3g6m 
crystal structure of a chitinase CrChi1 from the nematophagous fungus Clonostachys rosea in complex with a potent inhibitor caffeine 3hu7 
Structural characterization and binding studies of a plant pathogenesis related protein heamanthin from haemanthus multiflorus reveal its dual inhibitory effects against xylanase and alpha-amylase 3ian 
Crystal structure of a chitinase from Lactococcus lactis subsp. lactis 3m7s 
Crystal structure of the complex of xylanase GH-11 and alpha amylase inhibitor protein with cellobiose at 2.4 A resolution 3mu7 
Crystal structure of the xylanase and alpha-amylase inhibitor protein (XAIP-II) from scadoxus multiflorus at 1.2 A resolution 3n11 
Crystal stricture of wild-type chitinase from Bacillus cereus NCTU2 3n12 
Crystal stricture of chitinase in complex with zinc atoms from Bacillus cereus NCTU2 3n13 
Crystal stricture of D143A chitinase in complex with NAG from Bacillus cereus NCTU2 3n15 
Crystal stricture of E145Q chitinase in complex with NAG from Bacillus cereus NCTU2 3n17 
Crystal stricture of E145Q/Y227F chitinase in complex with NAG from Bacillus cereus NCTU2 3n18 
Crystal stricture of E145G/Y227F chitinase in complex with NAG from Bacillus cereus NCTU2 3n1a 
Crystal stricture of E145G/Y227F chitinase in complex with cyclo-(L-His-L-Pro) from Bacillus cereus NCTU2 3o9n 
Crystal Structure of a new form of xylanase-A-amylase inhibitor protein(XAIP-III) at 2.4 A resolution 3oa5 
The structure of chi1, a chitinase from Yersinia entomophaga 3oih 
Crystal Structure of the complex of xylanase-alpha-amylase inhibitor Protein (XAIP-I) with trehalose at 1.87 A resolution 3qok 
Crystal structure of putative chitinase II from Klebsiella pneumoniae 3rm4 
AMCase in complex with Compound 1 3rm8 
AMCase in complex with Compound 2 3rm9 
AMCase in complex with Compound 3 3rme 
AMCase in complex with Compound 5 3sim 
Crystallographic structure analysis of family 18 Chitinase from Crocus vernus 3w4r 
Crystal structure of an insect chitinase from the Asian corn borer, Ostrinia furnacalis 3wd0 
Serratia marcescens Chitinase B, tetragonal form 3wd1 
Serratia marcescens Chitinase B complexed with syn-triazole inhibitor 3wd2 
Serratia marcescens Chitinase B complexed with azide inhibitor 3wd3 
Serratia marcescens Chitinase B complexed with azide inhibitor 3wd4 
Serratia marcescens Chitinase B complexed with azide inhibitor and quinoline compound 3wij 
3WIJ 3wkz 
Crystal Structure of the Ostrinia furnacalis Group I Chitinase catalytic domain E148Q mutant 3wl0 
Crystal structure of Ostrinia furnacalis Group I chitinase catalytic domain E148A mutant in complex with a(GlcNAc)2 3wl1 
Crystal structure of Ostrinia furnacalis Group I chitinase catalytic domain in complex with reaction products (GlcNAc)2,3 3wqv 
3WQV 3wqw 
3WQW 4a5q 
Crystal structure of the chitinase Chi1 fitted into the 3D structure of the Yersinia entomophaga toxin complex 4ac1 
The structure of a fungal endo-beta-N-acetylglucosaminidase from glycosyl hydrolase family 18, at 1.3A resolution 4axn 
Hallmarks of processive and non-processive glycoside hydrolases revealed from computational and crystallographic studies of the Serratia marcescens chitinases 4ay1 
Human YKL-39 is a pseudo-chitinase with retained chitooligosaccharide binding properties 4dws 
Crystal Structure of a chitinase from the Yersinia entomophaga toxin complex 4hmc 
Crystal structure of cold-adapted chitinase from Moritella marina 4hmd 
Crystal structure of cold-adapted chitinase from Moritella marina with a reaction intermediate - oxazolinium ion (NGO) 4hme 
Crystal structure of cold-adapted chitinase from Moritella marina with a reaction product - NAG2 4lgx 
Structure of Chitinase D from Serratia proteamaculans revealed an unusually constrained substrate binding site 4mav 
Crystal structure of signaling protein SPB-40 complexed with 5-hydroxymethyl oxalanetriol at 2.80 A resolution 4mb3 
Crystal structure of E153Q mutant of cold-adapted chitinase from Moritella marina 4mb4 
Crystal structure of E153Q mutant of cold-adapted chitinase from Moritella complex with Nag4 4mb5 
Crystal structure of E153Q mutant of cold-adapted chitinase from Moritella complex with Nag5 4ml4 
Crystal structure of the complex of signaling glycoprotein from buffalo (SPB-40) with tetrahydropyran at 2.5 A resolution 4mnj 
4MNJ 4mnk 
4MNK 4mnl 
4MNL 4mnm 
4MNM 4mpk 
Crystal structure of the complex of buffalo signaling protein SPB-40 with N-acetylglucosamine at 2.65 A resolution 4mtv 
Crystal structure of the complex of Buffalo Signalling Glycoprotein with pentasaccharide at 2.8A resolution 4n42 
Crystal structure of allergen protein scam1 from Scadoxus multiflorus 4nsb 
Crystal structure of the complex of signaling glycoprotein, SPB-40 and N-acetyl salicylic acid at 3.05 A resolution 4nzc 
Crystal structure of Chitinase D from Serratia proteamaculans at 1.45 Angstrom resolution 4p8u 
4P8U 4p8v 
4P8V 4p8w 
4P8W 4p8x 
4P8X 4ptm 
Crystal Structure of Chitinase D from Serratia proteamaculans in complex with N-acetyl glucosamine, a hydrolyzed product of hexasaccharide at 1.7 Angstrom resolution 4q22 
Crystal structure of Chitinase D from Serratia proteamaculans in complex with N-acetyl glucosamine at 1.93 Angstrom resolution 4q6t 
The crystal structure of a class V chitininase from Pseudomonas fluorescens Pf-5 4q7n 
Crystal structure of the complex of Buffalo Signalling protein SPB-40 with 4-N-trimethylaminobutyraldehyde at 1.79 Angstrom Resolution 4r5e 
4R5E 4rl3 
4RL3 4s06 
4S06 4s19 
4S19 4s3j 
4S3J 4s3k 
4S3K 4toq 
4TOQ 4tx6 
4TX6 4tx8 
4TX8 4txe 
4TXE 4txg 
4TXG 4uri 
4URI 4w5u 
4W5U 4w5z 
4W5Z 4wiw 
4WIW 4wjx 
4WJX 4wk9 
4WK9 4wka 
4WKA 4wkf 
4WKF 4wkh 
4WKH 4z2g 
4Z2G 4z2h 
4Z2H 4z2i 
4Z2I 4z2j 
4Z2J 4z2k 
4Z2K 4z2l 
4Z2L 5dez 
5DEZ 5df0 
5DF0 5hbf 
5HBF 5jh8 
5JH8 5kz6 
5KZ6 - Links (links to other resources describing this domain)
-
INTERPRO IPR011583 PFAM Glyco_hydro_18

