The domain within your query sequence starts at position 2271 and ends at position 2350; the E-value for the IG domain shown below is 2.48e-8.
LSAMVAEEGQEATFQCVVSPSDAGVTWYKDGMQLQPSEKFVMVESGASRSLTILGLTLED AGQVTVEAEGASSSAALRVR
IGImmunoglobulin |
|---|
| SMART accession number: | SM00409 |
|---|---|
| Description: | - |
| Interpro abstract (IPR003599): | The basic structure of immunoglobulin (Ig) molecules is a tetramer of two light chains and two heavy chains linked by disulphide bonds. There are two types of light chains: kappa and lambda, each composed of a constant domain (CL) and a variable domain (VL). There are five types of heavy chains: alpha, delta, epsilon, gamma and mu, all consisting of a variable domain (VH) and three (in alpha, delta and gamma) or four (in epsilon and mu) constant domains (CH1 to CH4). Ig molecules are highly modular proteins, in which the variable and constant domains have clear, conserved sequence patterns. The domains in Ig and Ig-like molecules are grouped into four types: V-set (variable; IPR013106 ), C1-set (constant-1; IPR003597 ), C2-set (constant-2; IPR008424 ) and I-set (intermediate; IPR013098 ) [ (PUBMED:9417933) ]. Structural studies have shown that these domains share a common core Greek-key beta-sandwich structure, with the types differing in the number of strands in the beta-sheets as well as in their sequence patterns [ (PUBMED:15327963) (PUBMED:11377196) ]. Immunoglobulin-like domains that are related in both sequence and structure can be found in several diverse protein families. Ig-like domains are involved in a variety of functions, including cell-cell recognition, cell-surface receptors, muscle structure and the immune system [ (PUBMED:10698639) ]. This subfamily includes:
|
| Family alignment: |
There are 417572 IG domains in 173507 proteins in SMART's nrdb database.
Click on the following links for more information.
- Evolution (species in which this domain is found)
-
Taxonomic distribution of proteins containing IG domain.
This tree includes only several representative species. The complete taxonomic breakdown of all proteins with IG domain is also avaliable.
Click on the protein counts, or double click on taxonomic names to display all proteins containing IG domain in the selected taxonomic class.
- Disease (disease genes where sequence variants are found in this domain)
-
SwissProt sequences and OMIM curated human diseases associated with missense mutations within the IG domain.
Protein Disease Neural cell adhesion molecule L1 (P32004) (SMART) OMIM:308840: Hydrocephalus due to aqueductal stenosis
OMIM:307000: MASA syndrome
OMIM:303350: Spastic paraplegia
OMIM:312900:Low affinity immunoglobulin gamma Fc region receptor II-a (P12318) (SMART) OMIM:146790: {Lupus nephritis, susceptibility to} Myosin-binding protein C, cardiac-type (Q14896) (SMART) OMIM:600958: Cardiomyopathy, familial hypertrophic, 4
OMIM:115197:Netrin receptor DCC (P43146) (SMART) OMIM:120470: Colorectal cancer Poliovirus receptor (P15151) (SMART) OMIM:173850: {Polio, susceptibility to} Intercellular adhesion molecule 1 (P05362) (SMART) OMIM:147840: {Malaria, cerebral, susceptibility to} T-cell surface glycoprotein CD4 (P01730) (SMART) OMIM:186940: {Lupus erythematosus, susceptibility to} Myelin protein P0 (P25189) (SMART) OMIM:159440: Charcot-Marie-Tooth neuropathy-1B
OMIM:118200: Dejerine-Sottas disease, myelin P-zero-related
OMIM:145900: Hypomyelination, congenitalHigh affinity nerve growth factor receptor (P04629) (SMART) OMIM:191315: Insensitivity to pain, congenital, with anhidrosis
OMIM:256800: Medullary thyroid carcinoma, familial
OMIM:155240:Fibroblast growth factor receptor 2 (P21802) (SMART) OMIM:176943: Crouzon syndrome
OMIM:123500: Jackson-Weiss syndrome
OMIM:123150: Beare-Stevenson cutis gyrata syndrome
OMIM:123790: Pfeiffer syndrome
OMIM:101600: Apert syndrome
OMIM:101200: Saethre-Chotzen syndromeLow affinity immunoglobulin gamma Fc region receptor III-A (P08637) (SMART) OMIM:146740: {Lupus erythematosus, systemic, susceptibility}
OMIM:152700: Neutropenia, alloimmune neonatal ; {Viral infections, recurrent} - Metabolism (metabolic pathways involving proteins which contain this domain)
-
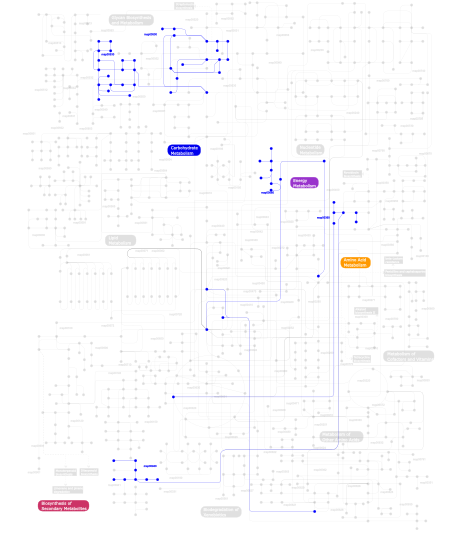
Click the image to view the interactive version of the map in iPath% proteins involved KEGG pathway ID Description 22.54 map04514 Cell adhesion molecules (CAMs) 9.23 map04060 Cytokine-cytokine receptor interaction 8.07 map04640 Hematopoietic cell lineage 7.72 map04360 Axon guidance 5.50 map04662 B cell receptor signaling pathway 4.79 map04650 Natural killer cell mediated cytotoxicity 4.44 map04670 Leukocyte transendothelial migration 4.35 map04510 Focal adhesion 3.11 map04520 Adherens junction 2.84 map04010 MAPK signaling pathway 2.66 map04660 T cell receptor signaling pathway 2.40 map04810 Regulation of actin cytoskeleton 2.31 map04612 Antigen processing and presentation 2.04 map04020 Calcium signaling pathway 1.77 map05120 Epithelial cell signaling in Helicobacter pylori infection 1.69 map05210 Colorectal cancer 1.69 map04210 Apoptosis 1.33 map05215 Prostate cancer 1.33 map05221 Acute myeloid leukemia 1.33 map05218 Melanoma 1.24 map05214 Glioma 1.24 map04540 Gap junction 0.98 map04940 Type I diabetes mellitus 0.89 map04916 Melanogenesis 0.80 map04370 VEGF signaling pathway 0.80 map04620 Toll-like receptor signaling pathway 0.62 map04664 Fc epsilon RI signaling pathway 0.53 map05216 Thyroid cancer 0.44  map00530
map00530Aminosugars metabolism 0.27 map05219 Bladder cancer 0.27 map04530 Tight junction 0.27 map04910 Insulin signaling pathway 0.18 map04512 ECM-receptor interaction 0.09  map00940
map00940Phenylpropanoid biosynthesis 0.09  map00680
map00680Methane metabolism 0.09  map00360
map00360Phenylalanine metabolism 0.09  map00500
map00500Starch and sucrose metabolism This information is based on mapping of SMART genomic protein database to KEGG orthologous groups. Percentage points are related to the number of proteins with IG domain which could be assigned to a KEGG orthologous group, and not all proteins containing IG domain. Please note that proteins can be included in multiple pathways, ie. the numbers above will not always add up to 100%.
- Structure (3D structures containing this domain)
3D Structures of IG domains in PDB
PDB code Main view Title 1a64 
ENGINEERING A MISFOLDED FORM OF RAT CD2 1a7b 
ENGINEERING A MISFOLDED FORM OF CD2 1ac6 
CRYSTAL STRUCTURE OF A VARIABLE DOMAIN MUTANT OF A T-CELL RECEPTOR ALPHA CHAIN 1b6u 
CRYSTAL STRUCTURE OF THE HUMAN KILLER CELL INHIBITORY RECEPTOR (KIR2DL3) SPECIFIC FOR HLA-CW3 RELATED ALLELES 1bd2 
COMPLEX BETWEEN HUMAN T-CELL RECEPTOR B7, VIRAL PEPTIDE (TAX) AND MHC CLASS I MOLECULE HLA-A 0201 1bqh 
MURINE CD8AA ECTODOMAIN FRAGMENT IN COMPLEX WITH H-2KB/VSV8 1bqs 
THE CRYSTAL STRUCTURE OF MUCOSAL ADDRESSIN CELL ADHESION MOLECULE-1 (MADCAM-1) 1ccz 
CRYSTAL STRUCTURE OF THE CD2-BINDING DOMAIN OF CD58 (LYMPHOCYTE FUNCTION-ASSOCIATED ANTIGEN 3) AT 1.8-A RESOLUTION 1cdc 
CD2, N-TERMINAL DOMAIN (1-99), TRUNCATED FORM 1ci5 
GLYCAN-FREE MUTANT ADHESION DOMAIN OF HUMAN CD58 (LFA-3) 1cid 
CRYSTAL STRUCTURE OF DOMAINS 3 & 4 OF RAT CD4 AND THEIR RELATIONSHIP TO THE NH2-TERMINAL DOMAINS 1dqt 
THE CRYSTAL STRUCTURE OF MURINE CTLA4 (CD152) 1dr9 
CRYSTAL STRUCTURE OF A SOLUBLE FORM OF B7-1 (CD80) 1e4j 
Crystal structure of soluble human Fc-gamma Receptor III (CD16) 1e4k 
Crystal structure of soluble human IgG1 Fc fragment-Fc-gamma Receptor III complex 1eaj 
DIMERIC STRUCTURE OF THE COXSACKIE VIRUS AND ADENOVIRUS RECEPTOR D1 DOMAIN AT 1.35 ANGSTROM RESOLUTION 1efx 
STRUCTURE OF A COMPLEX BETWEEN THE HUMAN NATURAL KILLER CELL RECEPTOR KIR2DL2 AND A CLASS I MHC LIGAND HLA-CW3 1f5w 
DIMERIC STRUCTURE OF THE COXSACKIE VIRUS AND ADENOVIRUS RECEPTOR D1 DOMAIN 1fcg 
ECTODOMAIN OF HUMAN FC GAMMA RECEPTOR, FCGRIIA 1flt 
VEGF IN COMPLEX WITH DOMAIN 2 OF THE FLT-1 RECEPTOR 1fnl 
CRYSTAL STRUCTURE OF THE EXTRACELLULAR DOMAIN OF A HUMAN FCGRIII 1fo0 
MURINE ALLOREACTIVE SCFV TCR-PEPTIDE-MHC CLASS I MOLECULE COMPLEX 1fyt 
CRYSTAL STRUCTURE OF A COMPLEX OF A HUMAN ALPHA/BETA-T CELL RECEPTOR, INFLUENZA HA ANTIGEN PEPTIDE, AND MHC CLASS II MOLECULE, HLA-DR1 1g0x 
CRYSTAL STRUCTURE OF THE LIGAND BINDING DOMAIN OF LIR-1 (ILT2) 1g0y 
IL-1 RECEPTOR TYPE 1 COMPLEXED WITH ANTAGONIST PEPTIDE AF10847 1g1c 
I1 DOMAIN FROM TITIN 1gsm 
A reassessment of the MAdCAM-1 structure and its role in integrin recognition. 1gxe 
central domain of cardiac myosin binding protein C 1h9v 
Human Fc-gamma-Receptor IIa (FcgRIIa), monoclinic 1hkf 
The three dimensional structure of NK cell receptor Nkp44, a triggering partner in natural cytotoxicity 1hng 
CRYSTAL STRUCTURE AT 2.8 ANGSTROMS RESOLUTION OF A SOLUBLE FORM OF THE CELL ADHESION MOLECULE CD2 1hxm 
Crystal Structure of a Human Vgamma9/Vdelta2 T Cell Receptor 1i8l 
HUMAN B7-1/CTLA-4 CO-STIMULATORY COMPLEX 1ij9 
Highly Hydrated Human VCAM-1 Fragment 1im9 
Crystal structure of the human natural killer cell inhibitory receptor KIR2DL1 bound to its MHC ligand HLA-Cw4 1ira 
COMPLEX OF THE INTERLEUKIN-1 RECEPTOR WITH THE INTERLEUKIN-1 RECEPTOR ANTAGONIST (IL1RA) 1itb 
TYPE-1 INTERLEUKIN-1 RECEPTOR COMPLEXED WITH INTERLEUKIN-1 BETA 1j8h 
Crystal Structure of a Complex of a Human alpha/beta-T cell Receptor, Influenza HA Antigen Peptide, and MHC Class II Molecule, HLA-DR4 1jew 
CRYO-EM STRUCTURE OF COXSACKIEVIRUS B3(M STRAIN) WITH ITS CELLULAR RECEPTOR, COXSACKIEVIRUS AND ADENOVIRUS RECEPTOR (CAR). 1kac 
KNOB DOMAIN FROM ADENOVIRUS SEROTYPE 12 IN COMPLEX WITH DOMAIN 1 OF ITS CELLULAR RECEPTOR CAR 1kb5 
MURINE T-CELL RECEPTOR VARIABLE DOMAIN/FAB COMPLEX 1kj2 
Murine Alloreactive ScFv TCR-Peptide-MHC Class I Molecule Complex 1lp9 
Xenoreactive complex AHIII 12.2 TCR bound to p1049/HLA-A2.1 1m4k 
Crystal structure of the human natural killer cell activator receptor KIR2DS2 (CD158j) 1nam 
MURINE ALLOREACTIVE SCFV TCR-PEPTIDE-MHC CLASS I MOLECULE COMPLEX 1nez 
The Crystal Structure of a TL/CD8aa Complex at 2.1A resolution:Implications for Memory T cell Generation, Co-receptor Preference and Affinity 1nfd 
AN ALPHA-BETA T CELL RECEPTOR (TCR) HETERODIMER IN COMPLEX WITH AN ANTI-TCR FAB FRAGMENT DERIVED FROM A MITOGENIC ANTIBODY 1nko 
Energetic and structural basis of sialylated oligosaccharide recognition by the natural killer cell inhibitory receptor p75/AIRM1 or Siglec-7 1nkr 
INHIBITORY RECEPTOR (P58-CL42) FOR HUMAN NATURAL KILLER CELLS 1npu 
CRYSTAL STRUCTURE OF THE EXTRACELLULAR DOMAIN OF MURINE PD-1 1o7s 
High resolution structure of Siglec-7 1o7v 
High resolution structure of Siglec-7 1od7 
N-terminal of Sialoadhesin in complex with Me-a-9-N-(naphthyl-2-carbonyl)-amino-9-deoxy-Neu5Ac (NAP compound) 1od9 
N-terminal of Sialoadhesin in complex with Me-a-9-N-benzoyl-amino-9-deoxy-Neu5Ac (BENZ compound) 1oda 
N-terminal of Sialoadhesin in complex with Me-a-9-N-(biphenyl-4-carbonyl)-amino-9-deoxy-Neu5Ac (BIP compound) 1oll 
Extracellular region of the human receptor NKp46 1olz 
The ligand-binding face of the semaphorins revealed by the high resolution crystal structure of SEMA4D 1ovz 
Crystal structure of human FcaRI 1ow0 
Crystal structure of human FcaRI bound to IgA1-Fc 1p53 
The Crystal Structure of ICAM-1 D3-D5 fragment 1p69 
STRUCTURAL BASIS FOR VARIATION IN ADENOVIRUS AFFINITY FOR THE CELLULAR RECEPTOR CAR (P417S MUTANT) 1p6a 
STRUCTURAL BASIS FOR VARIATION IN ASDENOVIRUS AFFINITY FOR THE CELLULAR RECEPTOR CAR (S489Y MUTANT) 1p6f 
Structure of the human natural cytotoxicity receptor NKp46 1p7q 
Crystal Structure of HLA-A2 Bound to LIR-1, a Host and Viral MHC Receptor 1pd6 
The NMR structure of domain C2 of human cardiac Myosin Binding Protein C 1q8m 
Crystal structure of the human myeloid cell activating receptor TREM-1 1qa9 
Structure of a Heterophilic Adhesion Complex Between the Human CD2 and CD58(LFA-3) Counter-Receptors 1qfo 
N-TERMINAL DOMAIN OF SIALOADHESIN (MOUSE) IN COMPLEX WITH 3'SIALYLLACTOSE 1qfp 
N-TERMINAL DOMAIN OF SIALOADHESIN (MOUSE) 1qsv 
THE VEGF-BINDING DOMAIN OF FLT-1, 20 NMR STRUCTURES 1qsz 
THE VEGF-BINDING DOMAIN OF FLT-1 (MINIMIZED MEAN) 1qty 
VASCULAR ENDOTHELIAL GROWTH FACTOR IN COMPLEX WITH DOMAIN 2 OF THE FLT-1 RECEPTOR 1rsf 
NMR Structure of Monomeric CAR d1 domain 1rv6 
Crystal Structure of PlGF in Complex with Domain 2 of VEGFR1 1smo 
Crystal Structure of Human Triggering Receptor Expressed on Myeloid Cells 1 (TREM-1) at 1.47 . 1sq2 
Crystal Structure Analysis of the Nurse Shark New Antigen Receptor (NAR) Variable Domain in Complex With Lysozyme 1t6v 
Crystal structure analysis of the nurse shark new antigen receptor (NAR) variable domain in complex with lysozyme 1t83 
CRYSTAL STRUCTURE OF A HUMAN TYPE III FC GAMMA RECEPTOR IN COMPLEX WITH AN FC FRAGMENT OF IGG1 (ORTHORHOMBIC) 1t89 
CRYSTAL STRUCTURE OF A HUMAN TYPE III FC GAMMA RECEPTOR IN COMPLEX WITH AN FC FRAGMENT OF IGG1 (HEXAGONAL) 1tit 
TITIN, IG REPEAT 27, NMR, MINIMIZED AVERAGE STRUCTURE 1tiu 
TITIN, IG REPEAT 27, NMR, 24 STRUCTURES 1u9k 
Crystal Structure of Mouse Triggering Receptor Expressed on Myeloid Cells 1 (TREM-1) at 1.76 1uct 
Crystal structure of the extracellular fragment of Fc alpha Receptor I (CD89) 1ufu 
Crystal structure of ligand binding domain of immunoglobulin-like transcript 2 (ILT2; LIR-1) 1ugn 
Crystal structure of LIR1.02, one of the alleles of LIR1 1url 
N-TERMINAL DOMAIN OF SIALOADHESIN (MOUSE) IN COMPLEX WITH GLYCOPEPTIDE 1vca 
CRYSTAL STRUCTURE OF AN INTEGRIN-BINDING FRAGMENT OF VASCULAR CELL ADHESION MOLECULE-1 AT 1.8 ANGSTROMS RESOLUTION 1vdg 
Crystal structure of LIR1.01, one of the alleles of LIR1 1ver 
Structure of New Antigen Receptor variable domain from sharks 1ves 
Structure of New Antigen Receptor variable domain from sharks 1vsc 
VCAM-1 1waa 
IG27 protein domain 1wit 
TWITCHIN IMMUNOGLOBULIN SUPERFAMILY DOMAIN (IGSF MODULE) (IG 18'), NMR, MINIMIZED AVERAGE STRUCTURE 1wiu 
TWITCHIN IMMUNOGLOBULIN SUPERFAMILY DOMAIN (IGSF MODULE) (IG 18'), NMR, 30 STRUCTURES 1x44 
Solution structure of the third ig-like domain of Myosin-dinding protein C, slow-type 1xau 
STRUCTURE OF THE BTLA ECTODOMAIN 1xed 
Crystal Structure of a Ligand-Binding Domain of the Human Polymeric Ig Receptor, pIgR 1xt5 
Crystal Structure of VCBP3, domain 1, from Branchiostoma floridae 1ypz 
Immune receptor 1z2k 
NMR structure of the D1 domain of the Natural Killer Cell Receptor, 2B4 1z7z 
Cryo-em structure of human coxsackievirus A21 complexed with five domain icam-1kilifi 1z9m 
Crystal Structure of Nectin-like molecule-1 protein Domain 1 1zgl 
Crystal structure of 3A6 TCR bound to MBP/HLA-DR2a 1zox 
CLM-1 Mouse Myeloid Receptor Extracellular Domain 2arj 
CD8alpha-alpha in complex with YTS 105.18 Fab 2atp 
Crystal structure of a CD8ab heterodimer 2avg 
NMR structure of cC1 domain from Human Cardiac Myosin Binding Protein C 2aw2 
Crystal structure of the human BTLA-HVEM complex 2bk8 
M1 domain from titin 2bve 
Structure of the N-terminal of Sialoadhesin in complex with 2-Phenyl- Prop5Ac 2c5d 
Structure of a minimal Gas6-Axl complex 2c9a 
Crystal structure of the MAM-Ig module of receptor protein tyrosine phosphatase mu 2coq 
Structure of new antigen receptor variable domain from sharks 2cqv 
Solution structure of the eighth Ig-like domain of human myosin light chain kinase 2cr6 
Solution structure of the Ig domain (2998-3100) of human obscurin 2cry 
Solution structure of the fifth ig-like domain of human kin of IRRE like 3 2d9c 
Solution structure of the first ig-like domain of signal-regulatory protein beta-1 (SIRP-beta-1) 2dav 
Solution structure of the first ig-like domain of Myosin-binding protein C, slow-type 2df3 
The structure of Siglec-7 in complex with alpha(2,3)/alpha(2,6) disialyl lactotetraosyl 2-(trimethylsilyl)ethyl 2dks 
Solution structure of the first IG-like domain of human carcinoembryonic antigen related cell adhesion molecule 8 2dku 
Solution structure of the third Ig-like domain of human KIAA1556 protein 2dl2 
KILLER IMMUNOGLOBULIN RECEPTOR 2DL2 2dli 
KILLER IMMUNOGLOBULIN RECEPTOR 2DL2,TRIGONAL FORM 2dlt 
Solution structure of the Ig-like domain(433- 525) of murine myosin-binding protein C, fast-type 2dru 
Crystal structure and binding properties of the CD2 and CD244 (2B4) binding protein, CD48 2dyp 
Crystal Structure of LILRB2(LIR2/ILT4/CD85d) complexed with HLA-G 2e5e 
Solution Structure of Variable-type Domain of Human Receptor for Advanced Glycation Endproducts 2e6p 
Solution structure of the Ig-like domain (714-804) from human Obscurin-like protein 1 2e7b 
Solution structure of the 6th Ig-like domain from human KIAA1556 2e7c 
Solution structure of the 6th Ig-like domain from human Myosin-binding protein C, fast-type 2edh 
Solution structure of the PDZ domain (3614- 3713 ) from human obscurin 2edk 
Solution structure of the third ig-like domain from human Myosin-binding protein C, fast-type 2edn 
Solution structure of the first ig-like domain from human Myosin-binding protein C, fast-type 2edo 
Solution structure of the first ig-like domain from human CD48 antigen 2edq 
Solution structure of the ig-like domain (3713-3806) of human obscurin 2edr 
Solution structure of the ig-like domain (3361-3449) of human obscurin 2edt 
Solution structure of the ig-like domain (3449-3537) from human Obscurin 2eny 
Solution structure of the ig-like domain (2735-2825) of human obscurin 2eo1 
Solution structure of the ig domain of human OBSCN protein 2esv 
Structure of the HLA-E-VMAPRTLIL/KK50.4 TCR complex 2fbo 
Crystal Structure of the Two Tandem V-type Regions of VCBP3 (v-region-containing chitin binding protein) to 1.85 A 2fcb 
HUMAN FC GAMMA RECEPTOR IIB ECTODOMAIN (CD32) 2frg 
Structure of the immunoglobulin-like domain of human TLT-1 2g5r 
Crystal structure of Siglec-7 in complex with methyl-9-(aminooxalyl-amino)-9-deoxyNeu5Ac (oxamido-Neu5Ac) 2gi7 
Crystal structure of human platelet Glycoprotein VI (GPVI) 2gk2 
Crystal structure of the N terminal domain of human CEACAM1 2gqh 
Solution structure of the 15th Ig-like domain of human KIAA1556 protein 2gw5 
Crystal Structure of LIR-2 (ILT4) at 1.8 : differences from LIR-1 (ILT2) in regions implicated in the binding of the Cytomegalovirus class I MHC homolog UL18 2hrl 
Siglec-7 in complex with GT1b 2i24 
Crystal structure analysis of the nurse shark New Antigen Receptor PBLA8 variable domain 2i25 
Crystal structure analysis of the nurse shark New antigen Receptor PBLA8 variable domain in complex with lysozyme 2i26 
Crystal structure analysis of the nurse shark new antigen receptor ancestral variable domain in complex with lysozyme 2i27 
Crystal Structure Analysis of the Nurse Shark New Antigen Receptor Ancestral variable domain 2ial 
Structural basis for recognition of mutant self by a tumor-specific, MHC class II-restricted TCR 2iam 
Structural basis for recognition of mutant self by a tumor-specific, MHC class II-restricted TCR 2ian 
Structural basis for recognition of mutant self by a tumor-specific, MHC class II-restricted TCR 2icc 
Extracellular Domain of CRIg 2ice 
CRIg bound to C3c 2icf 
CRIg bound to C3b 2if7 
Crystal Structure of NTB-A 2ifg 
Structure of the extracellular segment of human TRKA in complex with nerve growth factor 2ij0 
Structural basis of T cell specificity and activation by the bacterial superantigen toxic shock syndrome toxin-1 2ill 
Anomalous substructure of Titin-A168169 2j12 
Ad37 fibre head in complex with CAR D1 2j1k 
CAV-2 fibre head in complex with CAR D1 2j8h 
Structure of the immunoglobulin tandem repeat A168-A169 of titin 2j8o 
Structure of the immunoglobulin tandem repeat of titin A168-A169 2j8u 
Large CDR3a loop alteration as a function of MHC mutation. 2jcc 
AH3 recognition of mutant HLA-A2 W167A 2jjs 
Structure of human cd47 in complex with human signal regulatory protein (SIRP) alpha 2jjt 
Structure of human cd47 in complex with human signal regulatory protein (SIRP) alpha 2jju 
Structure of human signal regulatory protein (sirp) beta 2jjv 
Structure of human signal regulatory protein (sirp) beta(2) 2jjw 
Structure of human signal regulatory protein (sirp) gamma 2k1m 
3D NMR structure of domain cC0 of cardiac myosin binding protein C (MyBPC) 2kdg 
Solution Structure of the 1st Ig domain of Myotilin 2l7j 
Solution structure of the third Immunoglobulin-like domain of nectin-1 2l7u 
Structure of CEL-PEP-RAGE V domain complex 2lu7 
Solution NMR Structure of Ig like domain (1277-1357) of Obscurin-like protein 1 from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR8578D 2lvc 
Solution NMR Structure of Ig like domain (805-892) of Obscurin-like protein 1 from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR8578K 2m1k 
2M1K 2mjw 
2MJW 2mov 
2MOV 2mq0 
2MQ0 2mq3 
2MQ3 2mwc 
2MWC 2n56 
2N56 2n7a 
2N7A 2n7b 
2N7B 2nms 
The Crystal Structure of the Extracellular Domain of the Inhibitor Receptor Expressed on Myeloid Cells IREM-1 2nzi 
Crystal structure of domains A168-A170 from titin 2ocw 
Solution structure of human secretory component 2ol3 
crystal structure of BM3.3 ScFV TCR in complex with PBM8-H-2KBM8 MHC class I molecule 2or7 
Tim-2 2or8 
Tim-1 2otp 
Crystal Structure of Immunoglobulin-Like Transcript 1 (ILT1/LIR7/LILRA2) 2oyp 
T Cell Immunoglobulin Mucin-3 Crystal Structure Revealed a Galectin-9-independent Binding Surface 2oz4 
Structural Plasticity in IgSF Domain 4 of ICAM-1 Mediates Cell Surface Dimerization 2pet 
Lutheran glycoprotein, N-terminal domains 1 and 2. 2pf6 
Lutheran glycoprotein, N-terminal domains 1 and 2 2pkd 
Crystal structure of CD84: Insite into SLAM family function 2pnd 
Structure or murine CRIg 2ptt 
Structure of NK cell receptor 2B4 (CD244) bound to its ligand CD48 2ptu 
Structure of NK cell receptor 2B4 (CD244) 2ptv 
Structure of NK cell receptor ligand CD48 2pxy 
Crystal structures of immune receptor complexes 2q87 
The Crystal Structure of the Human IRp60 Ectodomain 2qhl 
Crystal Structure of Novel Immune-Type Receptor 10 Extracellular Fragment from Ictalurus punctatus 2qjd 
Crystal Structure of Novel Immune-Type Receptor 10 Extracellular Fragment Mutant N30D 2qqq 
Crystal Structure of Novel Immune-Type Receptor 11 Extracellular Fragment from Ictalurus punctatus 2qte 
Crystal Structure of Novel Immune-Type Receptor 11 Extracellular Fragment Mutant N30D 2rq8 
Solution NMR structure of titin I27 domain mutant 2uv3 
Structure of the signal-regulatory protein (SIRP) alpha domain that binds CD47. 2uwe 
Large CDR3a loop alteration as a function of MHC mutation 2v5y 
Crystal structure of the receptor protein tyrosine phosphatase mu ectodomain 2v6h 
Crystal structure of the C1 domain of cardiac myosin binding protein-C 2vsd 
crystal structure of CHIR-AB1 2w9l 
Canine adenovirus type 2 fibre head in complex with CAR domain D1 and sialic acid 2wbw 
Ad37 fibre head in complex with CAR D1 and sialic acid 2wp3 
Crystal structure of the Titin M10-Obscurin like 1 Ig complex 2wwk 
Crystal structure of the Titin M10-Obscurin like 1 Ig F17R mutant complex 2wwm 
Crystal structure of the Titin M10-Obscurin like 1 Ig complex in space group P1 2x1w 
Crystal Structure of VEGF-C in Complex with Domains 2 and 3 of VEGFR2 2x1x 
Crystal Structure of VEGF-C in Complex with Domains 2 and 3 of VEGFR2 in a Tetragonal Crystal Form 2xac 
Structural Insights into the Binding of VEGF-B by VEGFR-1D2: Recognition and Specificity 2xot 
Crystal structure of neuronal leucine rich repeat protein AMIGO-1 2y23 
CRYSTAL STRUCTURE OF THE MYOMESIN DOMAINS MY9-MY11 2y9r 
Crystal structure of the M10 domain of Titin 2yuv 
Solution Structure of 2nd Immunoglobulin Domain of Slow Type Myosin-Binding Protein C 2yuz 
Solution Structure of 4th Immunoglobulin Domain of Slow Type Myosin-Binding Protein C 2ywy 
Structure of new antigen receptor variable domain from sharks 2ywz 
Structure of new antigen receptor variable domain from sharks 2yxm 
Crystal structure of I-set domain of human Myosin Binding ProteinC 2yz1 
Crystal structure of the ligand-binding domain of murine SHPS-1/SIRP alpha 2yz8 
Crystal structure of the 32th Ig-like domain of human obscurin (KIAA1556) 2z31 
Crystal structure of immune receptor complex 2z35 
Crystal structure of immune receptor 2z8v 
Structure of an IgNAR-AMA1 complex 2z8w 
Structure of an IgNAR-AMA1 complex 2zg1 
Crystal Structure of Two N-terminal Domains of Siglec-5 in Complex with 6'-Sialyllactose 2zg2 
Crystal Structure of Two N-terminal Domains of Native Siglec-5 2zg3 
Crystal Structure of Two N-terminal Domains of Native Siglec-5 in Complex with 3'-Sialyllactose 2zwn 
Crystal structure of the novel two-domain type laccase from a metagenome 3arb 
Ternary crystal structure of the NKT TCR-CD1d-alpha-galactosylceramide analogue-OCH 3ard 
Ternary crystal structure of the mouse NKT TCR-CD1d-3'deoxy-alpha-galactosylceramide 3are 
Ternary crystal structure of the mouse NKT TCR-CD1d-4'deoxy-alpha-galactosylceramide 3arf 
Ternary crystal structure of the mouse NKT TCR-CD1d-C20:2 3arg 
Ternary crystal structure of the mouse NKT TCR-CD1d-alpha-glucosylceramide(C20:2) 3ay4 
Crystal structure of nonfucosylated Fc complexed with bis-glycosylated soluble form of Fc gamma receptor IIIa 3b5h 
Crystal structure of the extracellular portion of HAb18G/CD147 3b5t 
Crystal Structure of Novel Immune-Type Receptor 10 Se-Met Extracellular Fragment Mutant N30D 3b9k 
Crystal structure of CD8alpha-beta in complex with YTS 156.7 FAB 3bdb 
Crystal Structure of Novel Immune-Type Receptor 11 Extracellular Fragment from Ictalurus punctatus including Stalk Region 3bfo 
Crystal structure of Ig-like C2-type 2 domain of the human Mucosa-associated lymphoid tissue lymphoma translocation protein 1 3bi9 
Tim-4 3bia 
Tim-4 in complex with sodium potassium tartrate 3bib 
Tim-4 in complex with phosphatidylserine 3bik 
Crystal Structure of the PD-1/PD-L1 Complex 3bis 
Crystal Structure of the PD-L1 3bov 
Crystal structure of the receptor binding domain of mouse PD-L2 3bp5 
Crystal structure of the mouse PD-1 and PD-L2 complex 3bp6 
Crystal structure of the mouse PD-1 Mutant and PD-L2 complex 3c5z 
Crystal structure of mouse MHC class II I-Ab/3K peptide complexed with mouse TCR B3K506 3c60 
Crystal structure of mouse MHC class II I-Ab/3K peptide complexed with mouse TCR YAe62 3chn 
Solution structure of human secretory IgA1 3cjj 
Crystal structure of human rage ligand-binding domain 3cm9 
Solution Structure of Human SIgA2 3cx2 
Crystal structure of the C1 domain of cardiac isoform of myosin binding protein-C at 1.3A 3d2u 
Structure of UL18, a Peptide-Binding Viral MHC Mimic, Bound to a Host Inhibitory Receptor 3d5o 
Structural recognition and functional activation of FcrR by innate pentraxins 3dmm 
Crystal structure of the CD8 alpha beta/H-2Dd complex 3dx9 
Crystal Structure of the DM1 TCR at 2.75A 3dxa 
Crystal Structure of the DM1 TCR in complex with HLA-B*4405 and decamer EBV antigen 3fn3 
Dimeric Structure of PD-L1 3gsn 
Crystal structure of the public RA14 TCR in complex with the HCMV dominant NLV/HLA-A2 epitope 3h8n 
Crystal Structure Analysis of KIR2DS4 3he6 
Crystal structure of mouse CD1d-alpha-galactosylceramide with mouse Valpha14-Vbeta8.2 NKT TCR 3he7 
Crystal structure of mouse CD1d-alpha-galactosylceramide with mouse Valpha14-Vbeta7 NKT TCR 3j6l 
Kinetic and Structural Analysis of Coxsackievirus B3 Receptor Interactions and Formation of the A-particle 3j6m 
Kinetic and Structural Analysis of Coxsackievirus B3 Receptor Interactions and Formation of the A-particle 3kaa 
Structure of Tim-3 in complex with phosphatidylserine 3kg5 
Crystal structure of human Ig-beta homodimer 3kgr 
Crystal structure of the human leukocyte-associated Ig-like receptor-1 (LAIR-1) 3kho 
Crystal structure of murine Ig-beta (CD79b) homodimer 3khq 
Crystal structure of murine Ig-beta (CD79b) in the monomeric form 3knb 
Crystal structure of the titin C-terminus in complex with obscurin-like 1 3m45 
Crystal structure of Ig1 domain of mouse SynCAM 2 3mbe 
TCR 21.30 in complex with MHC class II I-Ag7HEL(11-27) 3mj6 
Crystal structure of the gammadelta T cell costimulatory receptor Junctional Adhesion Molecule-Like Protein, JAML 3mj7 
Crystal structure of the complex of JAML and Coxsackie and Adenovirus receptor, CAR 3mj9 
Crystal structure of JAML in complex with the stimulatory antibody HL4E10 3moq 
Amyloid beta(18-41) peptide fusion with new antigen receptor variable domain from sharks 3noi 
Crystal Structure of Natural Killer Cell Cytotoxicity Receptor NKp30 (NCR3) 3nvq 
Molecular mechanism of guidance cue recognition 3o3u 
Crystal Structure of Human Receptor for Advanced Glycation Endproducts (RAGE) 3o4l 
Genetic and structural basis for selection of a ubiquitous T cell receptor deployed in Epstein-Barr virus 3o4o 
Crystal structure of an Interleukin-1 receptor complex 3o8x 
Recognition of Glycolipid Antigen by iNKT Cell TCR 3o9w 
Recognition of a Glycolipid Antigen by the iNKT Cell TCR 3ol2 
Receptor-ligand structure of Human Semaphorin 4D with Plexin B1. 3oq3 
Structural Basis of Type-I Interferon Sequestration by a Poxvirus Decoy Receptor 3p2t 
Crystal Structure of Leukocyte Ig-like Receptor LILRB4 (ILT3/LIR-5/CD85k) 3pv6 
Crystal structure of NKp30 bound to its ligand B7-H6 3q0h 
Structure of T-cell immunoreceptor with immunoglobulin and ITIM domains (TIGIT) 3q2c 
Binding properties to HLA class I molecules and the structure of the leukocyte Ig-like receptor A3 (LILRA3/ILT6/LIR4/CD85e) 3q5o 
Crystal structure of human titin domain M10 3qdm 
The complex between TCR DMF4 and human Class I MHC HLA-A2 with the bound MART-1(26-35)(A27L) decameric peptide 3qeq 
The complex between TCR DMF4 and human Class I MHC HLA-A2 with the bound MART-1(27-35) nonameric peptide 3qi9 
Crystal structure of mouse CD1d-alpha-phosphotidylinositol with mouse Valpha14-Vbeta6 2A3-D NKT TCR 3qib 
Crystal structure of the 2B4 TCR in complex with MCC/I-Ek 3qiu 
Crystal structure of the 226 TCR in complex with MCC/I-Ek 3qiw 
Crystal structure of the 226 TCR in complex with MCC-p5E/I-Ek 3qjf 
Crystal structure of the 2B4 TCR 3qjh 
The crystal structure of the 5c.c7 TCR 3qp3 
Crystal structure of titin domain M4, tetragonal form 3qs7 
Crystal structure of a human Flt3 ligand-receptor ternary complex 3qs9 
Crystal structure of a human Flt3 ligand-receptor ternary complex 3qux 
Structure of the mouse CD1d-alpha-C-GalCer-iNKT TCR complex 3quy 
Structure of the mouse CD1d-BnNH-GSL-1'-iNKT TCR complex 3quz 
Structure of the mouse CD1d-NU-alpha-GalCer-iNKT TCR complex 3r0n 
Crystal Structure of the Immunoglobulin variable domain of Nectin-2 3r8b 
Crystal structure of Staphylococcal Enterotoxin B in complex with an affinity matured mouse TCR VBeta8.2 protein, G5-8 3rbg 
Crystal structure analysis of Class-I MHC restricted T-cell associated molecule 3rgv 
A single TCR bound to MHCI and MHC II reveals switchable TCR conformers 3rnk 
Crystal structure of the complex between mouse PD-1 mutant and PD-L2 IgV domain 3rnq 
Crystal structure of the complex between the extracellular domains of mouse PD-1 mutant and PD-L2 3rp1 
Crystal structure of Human LAIR-1 in C2 space group 3rq3 
Structure of T-cell immunoreceptor with immunoglobulin and ITIM domains (TIGIT) in hexagonal crystal form 3rtq 
Structure of the mouse CD1d-HS44-iNKT TCR complex 3ry4 
1.5 Angstrom resolution structure of glycosylated fcgammariia (low-responder polymorphism) 3ry5 
Three-dimensional structure of glycosylated fcgammariia (high-responder polymorphism) 3ry6 
Complex of fcgammariia (CD32) and the FC of human IGG1 3rzc 
Structure of the self-antigen iGb3 bound to mouse CD1d and in complex with the iNKT TCR 3s35 
Structural basis for the function of two anti-VEGF receptor antibodies 3s36 
Structural basis for the function of two anti-VEGF receptor antibodies 3s37 
Structural basis for the function of two anti-VEGF receptor antibodies 3sbw 
Crystal structure of the complex between the extracellular domains of mouse PD-1 mutant and human PD-L1 3scm 
Crystal structure of autoreactive-Valpha14-Vbeta6 NKT TCR in complex with CD1d-isoglobotrihexosylceramide 3sda 
Crystal structure of autoreactive-Valpha14-Vbeta6 NKT TCR in complex with CD1d-beta-galactosylceramide 3sdc 
Crystal structure of autoreactive-Valpha14-Vbeta6 NKT TCR in complex with CD1d-globotrihexosylceramide 3sdd 
Crystal structure of autoreactive-Valpha14-Vbeta6 NKT TCR in complex with CD1d-beta-lactosylceramide 3sgj 
Unique carbohydrate-carbohydrate interactions are required for high affinity binding between FcgIII and antibodies lacking core fucose 3sgk 
Unique carbohydrate/carbohydrate interactions are required for high affinity binding of FcgIII and antibodies lacking core fucose 3shs 
Three N-terminal domains of the bacteriophage RB49 Highly Immunogenic Outer Capsid protein (Hoc) 3ta3 
Structure of the mouse CD1d-Glc-DAG-s2-iNKT TCR complex 3tcx 
Structure of Engineered Single Domain ICAM-1 D1 with High-Affinity aL Integrin I Domain of Native C-Terminal Helix Conformation 3tn0 
Structure of mouse Va14Vb8.2NKT TCR-mouse CD1d-a-C-Galactosylceramide complex 3to4 
Structure of mouse Valpha14Vbeta2-mouseCD1d-alpha-Galactosylceramide 3tvm 
Structure of the mouse CD1d-SMC124-iNKT TCR complex 3ucr 
Crystal structure of the immunoreceptor TIGIT IgV domain 3udw 
Crystal structure of the immunoreceptor TIGIT in complex with Poliovirus receptor (PVR/CD155/necl-5) D1 domain 3vh8 
KIR3DL1 in complex with HLA-B*5701 3vxm 
The complex between C1-28 TCR and HLA-A24 bound to HIV-1 Nef134-10(2F) peptide 3wjj 
Crystal structure of IIb selective Fc variant, Fc(P238D), in complex with FcgRIIb 3wjl 
Crystal structure of IIb selective Fc variant, Fc(V12), in complex with FcgRIIb 3wn5 
3WN5 3wo3 
3WO3 3wo4 
3WO4 3wuw 
3WUW 3wuz 
3WUZ 3wv0 
3WV0 3wyr 
3WYR 4apq 
Crystal structure of autoreactive-Valpha14-Vbeta6 NKT TCR in complex with CD1d-sulfatide 4bfe 
Structure of the extracellular portion of mouse CD200RLa 4bfg 
Structure of the extracellular portion of mouse CD200R 4bfi 
Structure of the complex of the extracellular portions of mouse CD200R and mouse CD200 4bsj 
Crystal structure of VEGFR-3 extracellular domains D4-5 4bsk 
Crystal structure of VEGF-C in complex with VEGFR-3 domains D1-2 4c4k 
4C4K 4c56 
4C56 4ckv 
4CKV 4cl7 
4CL7 4cmm 
Structure of human CD47 in complex with human Signal Regulatory Protein (SIRP) alpha v1 4dep 
Structure of the IL-1b signaling complex 4dfh 
Crystal structure of cell adhesion molecule nectin-2/CD112 variable domain 4dfi 
Crystal structure of cell adhesion molecule nectin-2/CD112 mutant FAMP 4e41 
Structural basis for the recognition of mutant self by a tumor-specific, MHC class II-restricted T cell receptor G4 4e42 
Structural basis for the recognition of mutant self by a tumor-specific, MHC class II-restricted T cell receptor G4 4e9v 
Multicopper Oxidase mgLAC (data1) 4e9w 
Multicopper Oxidase mgLAC (data2) 4e9x 
Multicopper Oxidase mgLAC (data3) 4e9y 
Multicopper Oxidase mgLAC (data4) 4edq 
MBP-fusion protein of myosin-binding protein c residues 149-269 4f80 
Crystal Structure of the human BTN3A1 ectodomain 4f8q 
Crystal Structure of the Human BTN3A2 Ectodomain 4f8t 
Crystal Structure of the Human BTN3A3 Ectodomain 4f9l 
Crystal Structure of the Human BTN3A1 Ectodomain in Complex with the 20.1 Single Chain Antibody 4f9p 
Crystal Structure of the Human BTN3A1 Ectodomain in Complex with the 103.2 Single Chain Antibody 4fom 
Crystal structure of human nectin-3 full ectodomain (D1-D3) 4frw 
Crystal structure of human nectin-4 extracellular fragment D1-D2 4gaf 
Crystal structure of EBI-005, a chimera of human IL-1beta and IL-1Ra, bound to human Interleukin-1 receptor type 1 4gjt 
complex structure of nectin-4 bound to MV-H 4gkz 
HA1.7, a MHC class II restricted TCR specific for haemagglutinin 4gos 
Crystal structure of human B7-H4 IgV-like domain 4h1l 
TCR interaction with peptide mimics of nickel offers structural insights in nickel contact allergy 4h5s 
Complex structure of Necl-2 and CRTAM 4hbq 
Crystal structure of a loop deleted mutant of Human MAdCAM-1 D1D2 4hc1 
Crystal structure of a loop deleted mutant of human MAdCAM-1 D1D2 complexed with Fab 10G3 4hcr 
Crystal structure of human MAdCAM-1 D1D2 complexed with Fab PF-547659 4hd9 
Crystal structure of native human MAdCAM-1 D1D2 domain 4hgk 
Shark IgNAR variable domain 4hwn 
Crystal structure of the second Ig-C2 domain of human Fc-receptor like A (FCRLA), Isoform 9 [NYSGRC-005836] 4hza 
Crystal Structure of the Immunoglobulin variable domain of Nectin-2 in monoclinic form 4i0k 
Crystal structure of murine B7-H3 extracellular domain 4i9x 
Crystal structure of human cytomegalovirus glycoprotein UL141 targeting the death receptor TRAIL-R2 4im8 
low resolution crystal structure of mouse RAGE 4irj 
Structure of the mouse CD1d-4ClPhC-alpha-GalCer-iNKT TCR complex 4irs 
Structure of the mouse CD1d-PyrC-alpha-GalCer-iNKT TCR complex 4jgj 
Crystal structure of the Ig-like D1 domain from mouse Carcinoembryogenic antigen-related cell adhesion molecule 15 (CEACAM15) [PSI-NYSGRC-005691] 4jjh 
Crystal structure of the D1 domain from human Nectin-4 extracellular fragment [PSI-NYSGRC-005624] 4jkw 
Structure of the extracellular domain of butyrophilin BTN3A1 in complex with Isopentenyl pyrophosphate (IPP) 4jm0 
Structure of Human Cytomegalovirus Immune Modulator UL141 4jry 
Crystal Structure of SB47 TCR-HLA B*3505-LPEP complex 4k55 
Structure of the extracellular domain of butyrophilin BTN3A1 in complex with (E)-4-hydroxy-3-methyl-but-2-enyl pyrophosphate (HMBPP) 4k94 
Crystal structure of KIT D4D5 fragment in complex with anti-Kit antibody Fab19 4k9e 
Crystal structure of KIT D4D5 fragment in complex with anti-Kit antibodies Fab79D 4kjy 
Complex of high-affinity SIRP alpha variant FD6 with CD47 4kkn 
Crystal structure of bovine CTLA-4, PSI-NYSGRC-012704 4l1d 
Voltage-gated sodium channel beta3 subunit Ig domain 4ll9 
Crystal structure of D3D4 domain of the LILRB1 molecule 4lla 
Crystal structure of D3D4 domain of the LILRB2 molecule 4lp4 
Crystal structure of the human RAGE VC1 fragment in space group P62 4mnq 
TCR-peptide specificity overrides affinity enhancing TCR-MHC interactions 4mz2 
Crystal structure of the voltage-gated sodium channel beta 4 subunit extracellular domain 4mz3 
Crystal structure of the voltage-gated sodium channel beta 4 subunit extracellular domain, C131W mutant 4n8p 
Crystal structure of a strand swapped CTLA-4 from Duckbill Platypus [PSI-NYSGRC-012711] 4n8v 
Crystal structure of killer cell immunoglobulin-like receptor KIR2DS2 in complex with HLA-A 4nfb 
4NFB 4nfc 
4NFC 4nfd 
4NFD 4no0 
4NO0 4nob 
Crystal structure of the 1st Ig domain from mouse Polymeric Immunoglobulin receptor [PSI-NYSGRC-006220] 4nof 
Crystal structure of the second Ig domain from mouse Polymeric Immunoglobulin receptor [PSI-NYSGRC-006220] 4of0 
Crystal Structure of SYG-1 D1-D2, refolded 4of3 
Crystal Structure of SYG-1 D1-D2, Glycosylated 4of5 
Refinement of RAGE-DNA complex in 3S59 without DNA 4of6 
Crystal Structure of SYG-1 D1, Crystal form 1 4of7 
Crystal Structure of SYG-1 D1, Crystal Form 2 4of8 
Crystal Structure of Rst D1-D2 4ofd 
Crystal Structure of mouse Neph1 D1-D2 4ofi 
Crystal Structure of Duf (Kirre) D1 4ofv 
Refinement of RAGE-DNA complex in 3S58 without DNA 4ofy 
Crystal Structure of the Complex of SYG-1 D1-D2 and SYG-2 D1-D4 4oi7 
RAGE recognizes nucleic acids and promotes inflammatory responses to DNA 4oi8 
RAGE is a nucleic acid receptor that promotes inflammatory responses to DNA. 4oi9 
4OI9 4oia 
4OIA 4oib 
4OIB 4onh 
4ONH 4ozf 
JR5.1 protein complex 4ozg 
D2 protein complex 4ozh 
S16 protein complex 4p2o 
Crystal structure of the 2B4 TCR in complex with 2A/I-Ek 4p2q 
Crystal structure of the 5cc7 TCR in complex with 5c2/I-Ek 4p2r 
Crystal structure of the 5cc7 TCR in complex with 5c1/I-Ek 4p4k 
4P4K 4pbv 
4PBV 4pbw 
4PBW 4pgz 
4PGZ 4qeg 
4QEG 4qrp 
4QRP 4qxw 
4QXW 4r6u 
4R6U 4ra0 
4RA0 4rsv 
4RSV 4rwh 
4RWH 4u0q 
4U0Q 4udt 
4UDT 4udu 
4UDU 4uow 
4UOW 4v10 
4V10 4v2a 
4V2A 4whc 
4WHC 4whd 
4WHD 4wtz 
4WTZ 4wvr 
4WVR 4x5l 
4X5L 4y16 
4Y16 4y19 
4Y19 4y1a 
4Y1A 4y2d 
4Y2D 4y4f 
4Y4F 4y4h 
4Y4H 4y4k 
4Y4K 4y88 
4Y88 4y89 
4Y89 4y8a 
4Y8A 4yfd 
4YFD 4yiq 
4YIQ 4z18 
4Z18 4z7w 
4Z7W 4zak 
4ZAK 4zqk 
4ZQK 5a2f 
5A2F 5abd 
5ABD 5ayq 
5AYQ 5b38 
5B38 5b39 
5B39 5b5k 
5B5K 5bw7 
5BW7 5c14 
5C14 5c3t 
5C3T 5cj8 
5CJ8 5cjb 
5CJB 5d2l 
5D2L 5d4k 
5D4K 5d6d 
5D6D 5d7k 
5D7K 5d7l 
5D7L 5dzl 
5DZL 5dzn 
5DZN 5dzo 
5DZO 5e56 
5E56 5e5m 
5E5M 5e6i 
5E6I 5e9d 
5E9D 5eb9 
5EB9 5eiq 
5EIQ 5eiv 
5EIV 5f1s 
5F1S 5f4e 
5F4E 5f4t 
5F4T 5f4v 
5F4V 5f70 
5F70 5f71 
5F71 5f7f 
5F7F 5f7h 
5F7H 5fdy 
5FDY 5feb 
5FEB 5ffl 
5FFL 5fk9 
5FK9 5fka 
5FKA 5ggt 
5GGT 5grj 
5GRJ 5ius 
5IUS 5j89 
5J89 5j8o 
5J8O 5jdd 
5JDD 5jde 
5JDE 5jdj 
5JDJ 5jk9 
5JK9 5jkc 
5JKC 5jkd 
5JKD 5jke 
5JKE 5joe 
5JOE 5k6p 
5K6P 5l8j 
5L8J 5l8k 
5L8K 5l8l 
5L8L - Links (links to other resources describing this domain)
-
INTERPRO IPR003599 PFAM ig

