The domain within your query sequence starts at position 451 and ends at position 522; the E-value for the S1 domain shown below is 3.89e-20.
KIGTVVKGTVLAIKPFGILVKVGEQIKGLVPSMHLADIMMKNPEKKYSPGDEVKCRVLLC DPEAKKLIMTLK
S1Ribosomal protein S1-like RNA-binding domain |
|---|
| SMART accession number: | SM00316 |
|---|---|
| Description: | - |
| Interpro abstract (IPR022967): | The S1 domain was originally identified in ribosomal protein S1 but is found in a large number of RNA-associated proteins. The structure of the S1 RNA-binding domain from the Escherichia coli polynucleotide phosphorylase has been determined using NMR methods and consists of a five-stranded antiparallel beta barrel. Conserved residues on one face of the barrel and adjacent loops form the putative RNA-binding site [ (PUBMED:9008164) ]. The structure of the S1 domain is very similar to that of cold shock proteins. This suggests that they may both be derived from an ancient nucleic acid-binding protein [ (PUBMED:9008164) ]. |
| Family alignment: |
There are 254322 S1 domains in 154091 proteins in SMART's nrdb database.
Click on the following links for more information.
- Evolution (species in which this domain is found)
-
Taxonomic distribution of proteins containing S1 domain.
This tree includes only several representative species. The complete taxonomic breakdown of all proteins with S1 domain is also avaliable.
Click on the protein counts, or double click on taxonomic names to display all proteins containing S1 domain in the selected taxonomic class.
- Metabolism (metabolic pathways involving proteins which contain this domain)
-
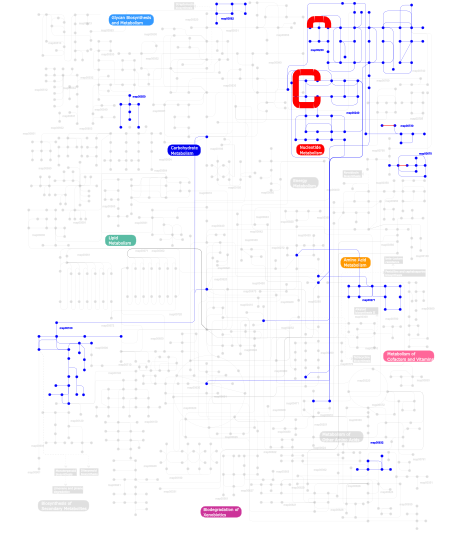
Click the image to view the interactive version of the map in iPath% proteins involved KEGG pathway ID Description 33.77  map00230
map00230Purine metabolism 33.60  map00240
map00240Pyrimidine metabolism 27.13 map03010 Ribosome 3.97 map03020 RNA polymerase 0.57  map00100
map00100Biosynthesis of steroids 0.40  map00550
map00550Peptidoglycan biosynthesis 0.08 map00970 Aminoacyl-tRNA biosynthesis 0.08 map02060 Phosphotransferase system (PTS) 0.08  map00562
map00562Inositol phosphate metabolism 0.08  map00670
map00670One carbon pool by folate 0.08  map00632
map00632Benzoate degradation via CoA ligation 0.08  map00730
map00730Thiamine metabolism 0.08  map00271
map00271Methionine metabolism This information is based on mapping of SMART genomic protein database to KEGG orthologous groups. Percentage points are related to the number of proteins with S1 domain which could be assigned to a KEGG orthologous group, and not all proteins containing S1 domain. Please note that proteins can be included in multiple pathways, ie. the numbers above will not always add up to 100%.
- Structure (3D structures containing this domain)
3D Structures of S1 domains in PDB
PDB code Main view Title 1ah9 
THE STRUCTURE OF THE TRANSLATIONAL INITIATION FACTOR IF1 FROM ESCHERICHIA COLI, NMR, 19 STRUCTURES 1e3h 
SeMet derivative of Streptomyces antibioticus PNPase/GPSI enzyme 1e3p 
tungstate derivative of Streptomyces antibioticus PNPase/GPSI enzyme 1go3 
Structure of an archeal homologue of the eukaryotic RNA polimerase II RPB4/RPB7 complex 1hh2 
Crystal structure of NusA from Thermotoga maritima 1hr0 
CRYSTAL STRUCTURE OF INITIATION FACTOR IF1 BOUND TO THE 30S RIBOSOMAL SUBUNIT 1k0r 
Crystal Structure of Mycobacterium tuberculosis NusA 1kl9 
Crystal structure of the N-terminal segment of Human eukaryotic initiation factor 2alpha 1l2f 
Crystal structure of NusA from Thermotoga maritima: a structure-based role of the N-terminal domain 1luz 
Crystal Structure of the K3L Protein From Vaccinia Virus (Wisconsin Strain) 1nt9 
Complete 12-subunit RNA polymerase II 1pqv 
RNA polymerase II-TFIIS complex 1q46 
crystal structure of the eIF2 alpha subunit from saccharomyces cerevisia 1q8k 
Solution structure of alpha subunit of human eIF2 1slj 
Solution structure of the S1 domain of RNase E from E. coli 1smx 
Crystal structure of the S1 domain of RNase E from E. coli (native) 1sn8 
Crystal structure of the S1 domain of RNase E from E. coli (Pb derivative) 1sro 
S1 RNA BINDING DOMAIN, NMR, 20 STRUCTURES 1wcm 
Complete 12-Subunit RNA Polymerase II at 3.8 A 1wi5 
Solution structure of the S1 RNA binding domain from human hypothetical protein BAA11502 1y14 
Crystal structure of yeast subcomplex of Rpb4 and Rpb7 1y1v 
Refined RNA Polymerase II-TFIIS complex 1y1w 
Complete RNA Polymerase II elongation complex 1y1y 
RNA Polymerase II-TFIIS-DNA/RNA complex 1y77 
Complete RNA Polymerase II elongation complex with substrate analogue GMPCPP 1yz6 
Crystal structure of intact alpha subunit of aIF2 from Pyrococcus abyssi 1zo1 
IF2, IF1, and tRNA fitted to cryo-EM data OF E. COLI 70S initiation complex 2a19 
PKR kinase domain- eIF2alpha- AMP-PNP complex. 2a1a 
PKR kinase domain-eIF2alpha Complex 2aho 
Structure of the archaeal initiation factor eIF2 alpha-gamma heterodimer from Sulfolobus solfataricus complexed with GDPNP 2asb 
Structure of a Mycobacterium tuberculosis NusA-RNA complex 2atw 
Structure of a Mycobacterium tuberculosis NusA-RNA complex 2b63 
Complete RNA Polymerase II-RNA inhibitor complex 2b8k 
12-subunit RNA Polymerase II 2ba0 
Archaeal exosome core 2ba1 
Archaeal exosome core 2bh8 
Combinatorial Protein 1b11 2bx2 
Catalytic domain of E. coli RNase E 2c0b 
Catalytic domain of E. coli RNase E in complex with 13-mer RNA 2c35 
Subunits Rpb4 and Rpb7 of human RNA polymerase II 2c4r 
Catalytic domain of E. coli RNase E 2cqo 
Solution structure of the S1 RNA binding domain of human hypothetical protein FLJ11067 2eqs 
Solution structure of the S1 RNA binding domain of human ATP-dependent RNA helicase DHX8 2id0 
Escherichia coli RNase II 2ix0 
RNase II 2ix1 
RNase II D209N mutant 2ja5 
CPD lesion containing RNA Polymerase II elongation complex A 2ja6 
CPD lesion containing RNA Polymerase II elongation complex B 2ja7 
CPD lesion containing RNA Polymerase II elongation complex C 2ja8 
CPD lesion containing RNA Polymerase II elongation complex D 2ja9 
Structure of the N-terminal deletion of yeast exosome component Rrp40 2je6 
Structure of a 9-subunit archaeal exosome 2jea 
Structure of a 9-subunit archaeal exosome bound to RNA 2jeb 
Structure of a 9-subunit archaeal exosome bound to Mn ions 2k4k 
Solution structure of GSP13 from Bacillus subtilis 2k52 
Structure of uncharacterized protein MJ1198 from Methanocaldococcus jannaschii. Northeast Structural Genomics Target MjR117B 2khi 
NMR structure of the domain 4 of the E. coli ribosomal protein S1 2khj 
NMR structure of the domain 6 of the E. coli ribosomal protein S1 2mfi 
Domain 1 of E. coli ribosomal protein S1 2mfl 
Domain 2 of E. coli ribosomal protein S1 2n3s 
2N3S 2n78 
2N78 2n8n 
2N8N 2nn6 
Structure of the human RNA exosome composed of Rrp41, Rrp45, Rrp46, Rrp43, Mtr3, Rrp42, Csl4, Rrp4, and Rrp40 2oce 
Crystal structure of Tex family protein PA5201 from Pseudomonas aeruginosa 2pmz 
Archaeal RNA polymerase from Sulfolobus solfataricus 2r7z 
Cisplatin lesion containing RNA polymerase II elongation complex 2r92 
Elongation complex of RNA polymerase II with artificial RdRP scaffold 2r93 
Elongation complex of RNA polymerase II with a hepatitis delta virus-derived RNA stem loop 2vmk 
Crystal Structure of E. coli RNase E Apoprotein - Catalytic Domain 2vnu 
Crystal structure of Sc Rrp44 2vrt 
Crystal Structure of E. coli RNase E possessing M1 RNA fragments - Catalytic Domain 2vum 
Alpha-amanitin inhibited complete RNA polymerase II elongation complex 2waq 
The complete structure of the archaeal 13-subunit DNA-directed RNA Polymerase 2wb1 
The complete structure of the archaeal 13-subunit DNA-directed RNA Polymerase 2wp8 
yeast rrp44 nuclease 2y0s 
Crystal structure of Sulfolobus shibatae RNA polymerase in P21 space group 2z0s 
Crystal structure of putative exosome complex RNA-binding protein 3aev 
Crystal structure of a/eIF2alpha-aDim2p-rRNA complex from Pyrococcus horikoshii OT3 3ayh 
Crystal structure of the C17/25 subcomplex from S. pombe RNA Polymerase III 3bzc 
Crystal Structure of the Tex protein from Pseudomonas aeruginosa, crystal form I 3bzk 
Crystal Structure of the Tex protein from Pseudomonas aeruginosa, crystal form 2 3cdi 
Crystal structure of E. coli PNPase 3cw2 
Crystal structure of the intact archaeal translation initiation factor 2 from Sulfolobus solfataricus . 3fki 
12-Subunit RNA Polymerase II Refined with Zn-SAD data 3go5 
Crystal structure of a multidomain protein with nucleic acid binding domains (sp_0946) from streptococcus pneumoniae tigr4 at 1.40 A resolution 3h0g 
RNA Polymerase II from Schizosaccharomyces pombe 3h3v 
Yeast RNAP II containing poly(A)-signal sequence in the active site 3hkz 
The X-ray crystal structure of RNA polymerase from Archaea 3hou 
Complete RNA polymerase II elongation complex I with a T-U mismatch 3hov 
Complete RNA polymerase II elongation complex II 3how 
Complete RNA polymerase II elongation complex III with a T-U mismatch and a frayed RNA 3'-uridine 3hox 
Complete RNA polymerase II elongation complex V 3hoy 
Complete RNA polymerase II elongation complex VI 3hoz 
Complete RNA polymerase II elongation complex IV with a T-U mismatch and a frayed RNA 3'-guanine 3i4m 
8-oxoguanine containing RNA polymerase II elongation complex D 3i4n 
8-oxoguanine containing RNA polymerase II elongation complex E 3i4o 
Crystal Structure of Translation Initiation Factor 1 from Mycobacterium tuberculosis 3j0k 
Orientation of RNA polymerase II within the human VP16-Mediator-pol II-TFIIF assembly 3j1n 
Cryo-EM map of a yeast minimal preinitiation complex interacting with the Mediator Head module 3j81 
3J81 3j9m 
3J9M 3jap 
3JAP 3jaq 
3JAQ 3jd5 
3JD5 3k1f 
Crystal structure of RNA Polymerase II in complex with TFIIB 3l7z 
Crystal structure of the S. solfataricus archaeal exosome 3m7n 
archaeoglobus fulgidus exosome with RNA bound to the active site 3m85 
Archaeoglobus fulgidus exosome y70a with RNA bound to the active site 3po2 
Arrested RNA Polymerase II elongation complex 3po3 
Arrested RNA Polymerase II reactivation intermediate 3qt1 
RNA polymerase II variant containing A Chimeric RPB9-C11 subunit 3v11 
Structure of the ternary initiation complex AIF2:GDPNP:methionylated initiator TRNA 4a3b 
RNA Polymerase II initial transcribing complex with a 4nt DNA-RNA hybrid 4a3c 
RNA Polymerase II initial transcribing complex with a 5nt DNA-RNA hybrid 4a3d 
RNA Polymerase II initial transcribing complex with a 6nt DNA-RNA hybrid 4a3e 
RNA Polymerase II initial transcribing complex with a 5nt DNA-RNA hybrid and soaked with AMPCPP 4a3f 
RNA Polymerase II initial transcribing complex with a 6nt DNA-RNA hybrid and soaked with AMPCPP 4a3g 
RNA Polymerase II initial transcribing complex with a 2nt DNA-RNA hybrid 4a3i 
RNA Polymerase II binary complex with DNA 4a3j 
RNA Polymerase II initial transcribing complex with a 2nt DNA-RNA hybrid and soaked with GMPCPP 4a3k 
RNA Polymerase II initial transcribing complex with a 7nt DNA-RNA hybrid 4a3l 
RNA Polymerase II initial transcribing complex with a 7nt DNA-RNA hybrid and soaked with AMPCPP 4a3m 
RNA Polymerase II initial transcribing complex with a 4nt DNA-RNA hybrid and soaked with AMPCPP 4a93 
RNA Polymerase II elongation complex containing a CPD Lesion 4aid 
Crystal structure of C. crescentus PNPase bound to RNase E recognition peptide 4aim 
Crystal structure of C. crescentus PNPase bound to RNase E recognition peptide 4am3 
Crystal structure of C. crescentus PNPase bound to RNA 4ayb 
RNAP at 3.2Ang 4ba1 
Archaeal exosome (Rrp4-Rrp41(D182A)-Rrp42) bound to inorganic phosphate 4ba2 
Archaeal exosome (Rrp4-Rrp41(D182A)-Rrp42) bound to inorganic phosphate 4bbr 
Structure of RNA polymerase II-TFIIB complex 4bbs 
Structure of an initially transcribing RNA polymerase II-TFIIB complex 4bxx 
Structures of RNA polymerase II complexes with Bye1, a chromatin- binding PHF3 DIDO homologue 4bxz 
RNA Polymerase II-Bye1 complex 4by1 
elongating RNA Polymerase II-Bye1 TLD complex soaked with AMPCPP 4by7 
elongating RNA Polymerase II-Bye1 TLD complex 4ifd 
Crystal structure of an 11-subunit eukaryotic exosome complex bound to RNA 4mtn 
Crystal structure of transcription termination factor NusA from Planctomyces limnophilus DSM 3776 4nbq 
4NBQ 4nng 
4NNG 4nnh 
4NNH 4nni 
4NNI 4nnk 
4NNK 4oo1 
4OO1 4oxp 
4OXP 4q7j 
4Q7J 4qiw 
4QIW 4qjf 
4QJF 4ql5 
4QL5 4r71 
4R71 4toi 
4TOI 4v1n 
4V1N 4v1o 
4V1O 4v8s 
4V8S 5aj3 
5AJ3 5aj4 
5AJ4 5c0w 
5C0W 5c0x 
5C0X 5c3e 
5C3E 5c44 
5C44 5c4a 
5C4A 5c4x 
5C4X 5c9s 
5C9S 5flm 
5FLM 5fmf 
5FMF 5fyw 
5FYW 5fz5 
5FZ5 5g06 
5G06 5ie8 
5IE8 5ip7 
5IP7 5ip9 
5IP9 5iy6 
5IY6 5iy7 
5IY7 5iy8 
5IY8 5iy9 
5IY9 5iya 
5IYA 5iyb 
5IYB 5iyc 
5IYC 5iyd 
5IYD 5jb3 
5JB3 5jea 
5JEA 5k0y 
5K0Y 5k36 
5K36 5lmn 
5LMN 5lmo 
5LMO 5lmp 
5LMP 5lmq 
5LMQ 5lmr 
5LMR 5lms 
5LMS 5lmt 
5LMT 5lmv 
5LMV 5sva 
5SVA - Links (links to other resources describing this domain)
-
PFAM S1 INTERPRO IPR022967

