The domain within your query sequence starts at position 412 and ends at position 491; the E-value for the S_TK_X domain shown below is 5.3e-9.
RSLDWQMVFLQKYPPPLIPPRGEVNAADAFDIGSFDEEDTKGIKLLDSDQELYRNFPLTI SERWQQEVAETVFDTINAET
S_TK_XExtension to Ser/Thr-type protein kinases |
|---|
| SMART accession number: | SM00133 |
|---|---|
| Description: | - |
| Interpro abstract (IPR000961): | Protein phosphorylation, which plays a key role in most cellular activities, is a reversible process mediated by protein kinases and phosphoprotein phosphatases. Protein kinases catalyse the transfer of the gamma phosphate from nucleotide triphosphates (often ATP) to one or more amino acid residues in a protein substrate side chain, resulting in a conformational change affecting protein function. Phosphoprotein phosphatases catalyse the reverse process. Protein kinases fall into three broad classes, characterised with respect to substrate specificity [ (PUBMED:3291115) ]:
Protein kinase function is evolutionarily conserved from Escherichia coli to human [ (PUBMED:12471243) ]. Protein kinases play a role in a multitude of cellular processes, including division, proliferation, apoptosis, and differentiation [ (PUBMED:12368087) ]. Phosphorylation usually results in a functional change of the target protein by changing enzyme activity, cellular location, or association with other proteins. The catalytic subunits of protein kinases are highly conserved, and several structures have been solved [ (PUBMED:15078142) ], leading to large screens to develop kinase-specific inhibitors for the treatments of a number of diseases [ (PUBMED:15320712) ]. The AGC (cAMP-dependent, cGMP-dependent and protein kinase C) protein kinase family embraces a collection of protein kinases that display a high degree of sequence similarity within their respective kinase domains. AGC kinase proteins are characterised by three conserved phosphorylation sites that critically regulate their function. The first one is located in an activation loop in the centre of the kinase domain. The two other phosphorylation sites are located outside the kinase domain in a conserved region on its C-terminal side, the AGC-kinase C-terminal domain. These sites serves as phosphorylation-regulated switches to control both intra- and inter-molecular interactions. Without these priming phosphorylations, the kinases are catalytically inactive [ (PUBMED:12495431) (PUBMED:11709088) (PUBMED:15209375) ]. Several structures of the AGC-kinase C-terminal domain have been solved. The first phosphorylation site is located in a turn motif, the second one at the end of the domain in an hydrophobic pocket. In PKB the phosphorylated hydrophobic motif engages a hydrophobic groove within the N-lobe of the kinase domain which orders alpha helices close to the active site [ (PUBMED:12434148) ]. |
| GO process: | protein phosphorylation (GO:0006468) |
| GO function: | protein serine/threonine kinase activity (GO:0004674), ATP binding (GO:0005524) |
| Family alignment: |
There are 38412 S_TK_X domains in 38361 proteins in SMART's nrdb database.
Click on the following links for more information.
- Evolution (species in which this domain is found)
-
Taxonomic distribution of proteins containing S_TK_X domain.
This tree includes only several representative species. The complete taxonomic breakdown of all proteins with S_TK_X domain is also avaliable.
Click on the protein counts, or double click on taxonomic names to display all proteins containing S_TK_X domain in the selected taxonomic class.
- Disease (disease genes where sequence variants are found in this domain)
-
SwissProt sequences and OMIM curated human diseases associated with missense mutations within the S_TK_X domain.
Protein Disease Rhodopsin kinase (Q15835) (SMART) OMIM:180381: Oguchi disease-2
OMIM:258100:Protein kinase C gamma type (P05129) (SMART) OMIM:176980: PROTEIN KINASE C, GAMMA; PRKCG - Metabolism (metabolic pathways involving proteins which contain this domain)
-
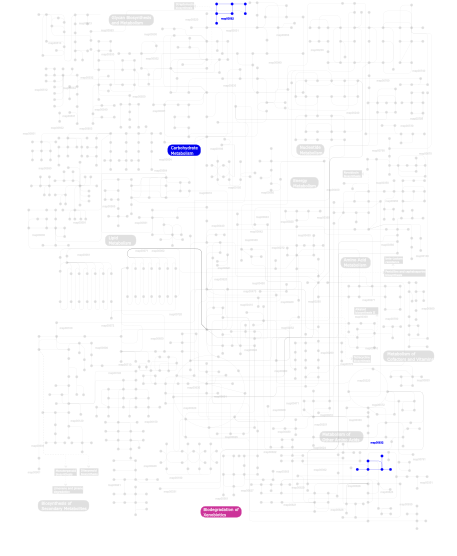
Click the image to view the interactive version of the map in iPath% proteins involved KEGG pathway ID Description 6.60 map04010 MAPK signaling pathway 4.74 map04720 Long-term potentiation 4.74 map04910 Insulin signaling pathway 4.64 map04530 Tight junction 4.49 map04914 Progesterone-mediated oocyte maturation 4.20 map04310 Wnt signaling pathway 4.20 map04540 Gap junction 3.42 map04510 Focal adhesion 3.42 map04012 ErbB signaling pathway 3.37 map04020 Calcium signaling pathway 3.37 map04916 Melanogenesis 3.37 map04150 mTOR signaling pathway 3.13 map04210 Apoptosis 2.78 map04740 Olfactory transduction 2.59 map04370 VEGF signaling pathway 2.59 map05214 Glioma 2.59 map05223 Non-small cell lung cancer 2.25 map04670 Leukocyte transendothelial migration 2.25 map04730 Long-term depression 2.00 map05221 Acute myeloid leukemia 1.95 map04742 Taste transduction 1.95 map04912 GnRH signaling pathway 1.95 map04340 Hedgehog signaling pathway 1.66 map04350 TGF-beta signaling pathway 1.42 map04070 Phosphatidylinositol signaling system 1.42 map04650 Natural killer cell mediated cytotoxicity 1.17 map05222 Small cell lung cancer 1.17 map05213 Endometrial cancer 1.17 map04662 B cell receptor signaling pathway 1.17 map05212 Pancreatic cancer 1.17 map04630 Jak-STAT signaling pathway 1.17 map05220 Chronic myeloid leukemia 1.17 map04920 Adipocytokine signaling pathway 1.17 map04664 Fc epsilon RI signaling pathway 1.17 map05210 Colorectal cancer 1.17 map04620 Toll-like receptor signaling pathway 1.17 map05215 Prostate cancer 1.17 map04660 T cell receptor signaling pathway 1.17 map05211 Renal cell carcinoma 1.17 map05218 Melanoma 0.83 map04810 Regulation of actin cytoskeleton 0.83 map04360 Axon guidance 0.29  map00632
map00632Benzoate degradation via CoA ligation 0.29  map00562
map00562Inositol phosphate metabolism 0.24 map04111 Cell cycle - yeast This information is based on mapping of SMART genomic protein database to KEGG orthologous groups. Percentage points are related to the number of proteins with S_TK_X domain which could be assigned to a KEGG orthologous group, and not all proteins containing S_TK_X domain. Please note that proteins can be included in multiple pathways, ie. the numbers above will not always add up to 100%.
- Structure (3D structures containing this domain)
3D Structures of S_TK_X domains in PDB
PDB code Main view Title 1apm 
2.0 ANGSTROM REFINED CRYSTAL STRUCTURE OF THE CATALYTIC SUBUNIT OF CAMP-DEPENDENT PROTEIN KINASE COMPLEXED WITH A PEPTIDE INHIBITOR AND DETERGENT 1atp 
2.2 ANGSTROM REFINED CRYSTAL STRUCTURE OF THE CATALYTIC SUBUNIT OF CAMP-DEPENDENT PROTEIN KINASE COMPLEXED WITH MNATP AND A PEPTIDE INHIBITOR 1bkx 
A BINARY COMPLEX OF THE CATALYTIC SUBUNIT OF CAMP-DEPENDENT PROTEIN KINASE AND ADENOSINE FURTHER DEFINES CONFORMATIONAL FLEXIBILITY 1bx6 
CRYSTAL STRUCTURE OF THE POTENT NATURAL PRODUCT INHIBITOR BALANOL IN COMPLEX WITH THE CATALYTIC SUBUNIT OF CAMP-DEPENDENT PROTEIN KINASE 1cdk 
CAMP-DEPENDENT PROTEIN KINASE CATALYTIC SUBUNIT (E.C.2.7.1.37) (PROTEIN KINASE A) COMPLEXED WITH PROTEIN KINASE INHIBITOR PEPTIDE FRAGMENT 5-24 (PKI(5-24) ISOELECTRIC VARIANT CA) AND MN2+ ADENYLYL IMIDODIPHOSPHATE (MNAMP-PNP) AT PH 5.6 AND 7C AND 4C 1cmk 
CRYSTAL STRUCTURES OF THE MYRISTYLATED CATALYTIC SUBUNIT OF CAMP-DEPENDENT PROTEIN KINASE REVEAL OPEN AND CLOSED CONFORMATIONS 1ctp 
STRUCTURE OF THE MAMMALIAN CATALYTIC SUBUNIT OF CAMP-DEPENDENT PROTEIN KINASE AND AN INHIBITOR PEPTIDE DISPLAYS AN OPEN CONFORMATION 1fmo 
CRYSTAL STRUCTURE OF A POLYHISTIDINE-TAGGED RECOMBINANT CATALYTIC SUBUNIT OF CAMP-DEPENDENT PROTEIN KINASE COMPLEXED WITH THE PEPTIDE INHIBITOR PKI(5-24) AND ADENOSINE 1fot 
STRUCTURE OF THE UNLIGANDED CAMP-DEPENDENT PROTEIN KINASE CATALYTIC SUBUNIT FROM SACCHAROMYCES CEREVISIAE 1gzk 
Molecular mechanism for the regulation of protein kinase B/Akt by hydrophobic motif phosphorylation 1gzn 
Structure of PKB kinase domain 1gzo 
Structure of protein kinase B unphosphorylated 1j3h 
Crystal structure of apoenzyme cAMP-dependent protein kinase catalytic subunit 1jbp 
Crystal Structure of the Catalytic Subunit of cAMP-dependent Protein Kinase Complexed with a Substrate Peptide, ADP and Detergent 1jlu 
Crystal Structure of the Catalytic Subunit of cAMP-dependent Protein Kinase Complexed with a Phosphorylated Substrate Peptide and Detergent 1l3r 
Crystal Structure of a Transition State Mimic of the Catalytic Subunit of cAMP-dependent Protein Kinase 1mrv 
crystal structure of an inactive Akt2 kinase domain 1mry 
crystal structure of an inactive akt2 kinase domain 1o6k 
Structure of activated form of PKB kinase domain S474D with GSK3 peptide and AMP-PNP 1o6l 
Crystal structure of an activated Akt/protein kinase B (PKB-PIF chimera) ternary complex with AMP-PNP and GSK3 peptide 1omw 
Crystal Structure of the complex between G Protein-Coupled Receptor Kinase 2 and Heterotrimeric G Protein beta 1 and gamma 2 subunits 1q24 
PKA double mutant model of PKB in complex with MgATP 1q61 
PKA triple mutant model of PKB 1q62 
PKA double mutant model of PKB 1q8t 
The Catalytic Subunit of cAMP-dependent Protein Kinase (PKA) in Complex with Rho-kinase Inhibitor Y-27632 1q8u 
The Catalytic Subunit of cAMP-dependent Protein Kinase in Complex with Rho-kinase Inhibitor H-1152P 1q8w 
The Catalytic Subunit of cAMP-dependent Protein Kinase in Complex with Rho-kinase Inhibitor Fasudil (HA-1077) 1rdq 
Hydrolysis of ATP in the crystal of Y204A mutant of cAMP-dependent protein kinase 1re8 
Crystal structure of cAMP-dependent protein kinase complexed with balanol analog 2 1rej 
Crystal structure of cAMP-dependent protein kinase complexed with balanol analog 1 1rek 
Crystal structure of cAMP-dependent protein kinase complexed with balanol analog 8 1smh 
Protein kinase A variant complex with completely ordered N-terminal helix 1stc 
CAMP-DEPENDENT PROTEIN KINASE, ALPHA-CATALYTIC SUBUNIT IN COMPLEX WITH STAUROSPORINE 1sve 
Crystal Structure of Protein Kinase A in Complex with Azepane Derivative 1 1svg 
Crystal Structure of Protein Kinase A in Complex with Azepane Derivative 4 1svh 
Crystal Structure of Protein Kinase A in Complex with Azepane Derivative 8 1syk 
Crystal structure of E230Q mutant of cAMP-dependent protein kinase reveals unexpected apoenzyme conformation 1szm 
DUAL BINDING MODE OF BISINDOLYLMALEIMIDE 2 TO PROTEIN KINASE A (PKA) 1veb 
Crystal Structure of Protein Kinase A in Complex with Azepane Derivative 5 1xh4 
Crystal Structures of Protein Kinase B Selective Inhibitors in Complex with Protein Kinase A and Mutants 1xh5 
Crystal Structures of Protein Kinase B Selective Inhibitors in Complex with Protein Kinase A and Mutants 1xh6 
Crystal Structures of Protein Kinase B Selective Inhibitors in Complex with Protein Kinase A and Mutants 1xh7 
Crystal Structures of Protein Kinase B Selective Inhibitors in Complex with Protein Kinase A and Mutants 1xh8 
Crystal Structures of Protein Kinase B Selective Inhibitors in Complex with Protein Kinase A and Mutants 1xh9 
Crystal Structures of Protein Kinase B Selective Inhibitors in Complex with Protein Kinase A and Mutants 1xha 
Crystal Structures of Protein Kinase B Selective Inhibitors in Complex with Protein Kinase A and Mutants 1xjd 
Crystal Structure of PKC-theta complexed with Staurosporine at 2A resolution 1ydr 
STRUCTURE OF CAMP-DEPENDENT PROTEIN KINASE, ALPHA-CATALYTIC SUBUNIT IN COMPLEX WITH H7 PROTEIN KINASE INHIBITOR 1-(5-ISOQUINOLINESULFONYL)-2-METHYLPIPERAZINE 1yds 
STRUCTURE OF CAMP-DEPENDENT PROTEIN KINASE, ALPHA-CATALYTIC SUBUNIT IN COMPLEX WITH H8 PROTEIN KINASE INHIBITOR [N-(2-METHYLAMINO)ETHYL]-5-ISOQUINOLINESULFONAMIDE 1ydt 
STRUCTURE OF CAMP-DEPENDENT PROTEIN KINASE, ALPHA-CATALYTIC SUBUNIT IN COMPLEX WITH H89 PROTEIN KINASE INHIBITOR N-[2-(4-BROMOCINNAMYLAMINO)ETHYL]-5-ISOQUINOLINE 1ym7 
G Protein-Coupled Receptor Kinase 2 (GRK2) 1zrz 
Crystal Structure of the Catalytic Domain of Atypical Protein Kinase C-iota 2acx 
Crystal Structure of G protein coupled receptor kinase 6 bound to AMPPNP 2bcj 
Crystal Structure of G Protein-Coupled Receptor Kinase 2 in Complex with Galpha-q and Gbetagamma Subunits 2c1a 
Structure of cAMP-dependent protein kinase complexed with Isoquinoline-5-sulfonic acid (2-(2-(4-chlorobenzyloxy)ethylamino) ethyl)amide 2c1b 
Structure of cAMP-dependent protein kinase complexed with (4R,2S)-5'-( 4-(4-Chlorobenzyloxy)pyrrolidin-2-ylmethanesulfonyl)isoquinoline 2cpk 
CRYSTAL STRUCTURE OF THE CATALYTIC SUBUNIT OF CYCLIC ADENOSINE MONOPHOSPHATE-DEPENDENT PROTEIN KINASE 2erz 
Crystal Structure of c-AMP Dependent Kinase (PKA) bound to hydroxyfasudil 2esm 
Crystal Structure of ROCK 1 bound to fasudil 2etk 
Crystal Structure of ROCK 1 bound to hydroxyfasudil 2etr 
Crystal Structure of ROCK I bound to Y-27632 2f2u 
crystal structure of the Rho-kinase kinase domain 2f7e 
PKA complexed with (S)-2-(1H-Indol-3-yl)-1-(5-isoquinolin-6-yl-pyridin-3-yloxymethyl-etylamine 2f7x 
Protein Kinase A bound to (S)-2-(1H-Indol-3-yl)-1-[5-((E)-2-pyridin-4-yl-vinyl)-pyridin-3-yloxymethyl]-ethylamine 2f7z 
Protein Kinase A bound to (R)-1-(1H-Indol-3-ylmethyl)-2-(2-pyridin-4-yl-[1,7]naphtyridin-5-yloxy)-ehylamine 2gfc 
cAMP-dependent protein kinase PKA catalytic subunit with PKI-5-24 2gnf 
Protein kinase A fivefold mutant model of Rho-kinase with Y-27632 2gng 
Protein kinase A fivefold mutant model of Rho-kinase 2gnh 
PKA five fold mutant model of Rho-kinase with H1152P 2gni 
PKA fivefold mutant model of Rho-kinase with inhibitor Fasudil (HA1077) 2gnj 
PKA three fold mutant model of Rho-kinase with Y-27632 2gnl 
PKA threefold mutant model of Rho-kinase with inhibitor H-1152P 2gu8 
Discovery of 2-Pyrimidyl-5-Amidothiophenes as Novel and Potent Inhibitors for AKT: Synthesis and SAR Studies 2h9v 
Structural basis for induced-fit binding of Rho-kinase to the inhibitor Y27632 2i0e 
Structure of catalytic domain of human protein kinase C beta II complexed with a bisindolylmaleimide inhibitor 2jdo 
STRUCTURE OF PKB-BETA (AKT2) COMPLEXED WITH ISOQUINOLINE-5-SULFONIC ACID (2-(2-(4-CHLOROBENZYLOXY) ETHYLAMINO)ETHYL)AMIDE 2jdr 
STRUCTURE OF PKB-BETA (AKT2) COMPLEXED WITH THE INHIBITOR A-443654 2jds 
Structure of cAMP-dependent protein kinase complexed with A-443654 2jdt 
Structure of PKA-PKB chimera complexed with ISOQUINOLINE-5-SULFONIC ACID (2-(2-(4-CHLOROBENZYLOXY) ETHYLAMINO)ETHYL)AMIDE 2jdv 
Structure of PKA-PKB chimera complexed with A-443654 2jed 
The crystal structure of the kinase domain of the protein kinase C theta in complex with NVP-XAA228 at 2.32A resolution. 2oh0 
Crystal structure of Protein Kinase A in complex with Pyridine-Pyrazolopyridine Based Inhibitors 2ojf 
Crystal structure of Protein Kinase A in complex with Pyridine-Pyrazolopyridine based inhibitors 2qcs 
A complex structure between the Catalytic and Regulatory subunit of Protein Kinase A that represents the inhibited state 2qur 
Crystal Structure of F327A/K285P Mutant of cAMP-dependent Protein Kinase 2qvs 
Crystal Structure of Type IIa Holoenzyme of cAMP-dependent Protein Kinase 2r5t 
Crystal Structure of Inactive Serum and Glucocorticoid- Regulated Kinase 1 in Complex with AMP-PNP 2uvx 
Structure of PKA-PKB chimera complexed with 7-azaindole 2uvy 
Structure of PKA-PKB chimera complexed with methyl-(4-(9H-purin-6-yl)- benzyl)-amine 2uvz 
Structure of PKA-PKB chimera complexed with C-Phenyl-C-(4-(9H-purin-6- yl)-phenyl)-methylamine 2uw0 
Structure of PKA-PKB chimera complexed with 6-(4-(4-(4-Chloro-phenyl) -piperidin-4-yl)-phenyl)-9H-purine 2uw3 
Structure of PKA-PKB chimera complexed with 5-methyl-4-phenyl-1H- pyrazole 2uw4 
Structure of PKA-PKB chimera complexed with 2-(4-(5-methyl-1H-pyrazol- 4-yl)-phenyl)-ethylamine 2uw5 
Structure of PKA-PKB chimera complexed with (R)-2-(4-chloro-phenyl)- 2-(4-1H-pyrazol-4-yl)-phenyl)-ethylamine 2uw6 
Structure of PKA-PKB chimera complexed with (S)-2-(4-chloro-phenyl)- 2-(4-1H-pyrazol-4-yl)-phenyl)-ethylamine 2uw7 
Structure of PKA-PKB chimera complexed with 4-(4-chloro-phenyl)-4-(4- (1H-pyrazol-4-yl)-phenyl)-piperidine 2uw8 
Structure of PKA-PKB chimera complexed with 2-(4-chloro-phenyl)-2- phenyl-ethylamine 2uw9 
STRUCTURE OF PKB-BETA (AKT2) COMPLEXED WITH 4-(4-chloro-phenyl)-4-(4-( 1H-pyrazol-4-yl)-phenyl)-piperidine 2uzt 
PKA structures of AKT, indazole-pyridine inhibitors 2uzu 
PKA structures of indazole-pyridine series of AKT inhibitors 2uzv 
PKA structures of indazole-pyridine series of AKT inhibitors 2uzw 
PKA structures of indazole-pyridine series of AKT inhibitors 2v55 
Mechanism of multi-site phosphorylation from a ROCK-I:RhoE complex structure 2vd5 
Structure of Human Myotonic Dystrophy Protein Kinase in Complex with the Bisindoylmaleide inhibitor BIM VIII 2vnw 
Structure of PKA-PKB chimera complexed with (1-(9H-Purin-6-yl) piperidin-4-yl)methanamine 2vny 
Structure of PKA-PKB chimera complexed with (1-(9H-Purin-6-yl) piperidin-4-yl)amine 2vo0 
Structure of PKA-PKB chimera complexed with C-(4-(4-Chlorophenyl)-1-( 7H-pyrrolo(2,3-d)pyrimidin-4-yl)piperidin-4-yl)methylamine 2vo3 
Structure of PKA-PKB chimera complexed with C-(4-(4-Chlorobenzyl)-1-( 7H-pyrrolo(2,3-d)pyrimidin-4-yl)piperidin-4-yl)methylamine 2vo6 
Structure of PKA-PKB chimera complexed with 4-(4-Chlorobenzyl)-1-(7H- pyrrolo(2,3-d)pyrimidin-4-yl)piperidin-4-ylamine 2vo7 
Structure of PKA complexed with 4-(4-Chlorobenzyl)-1-(7H-pyrrolo(2,3- d)pyrimidin-4-yl)piperidin-4-ylamine 2x39 
Structure of 4-Amino-N-(4-chlorobenzyl)-1-(7H-pyrrolo(2,3-d)pyrimidin- 4-yl)piperidine-4-carboxamide bound to PKB 2xh5 
Structure of 4-(4-tert-Butylbenzyl)-1-(7H-pyrrolo(2,3-d)pyrimidin-4- yl)piperidin-4-amine bound to PKB 2z7q 
Crystal structure of the N-terminal kinase domain of human RSK-1 bound to AMP-PCP 2z7r 
Crystal Structure of the N-terminal Kinase Domain of Human RSK1 bound to Staurosporine 2z7s 
Crystal Structure of the N-terminal Kinase Domain of Human RSK1 bound to Purvalnol A 3a60 
Crystal structure of unphosphorylated p70S6K1 (Form I) 3a61 
Crystal structure of unphosphorylated p70S6K1 (Form II) 3a62 
Crystal structure of phosphorylated p70S6K1 3a8w 
Crystal Structure of PKCiota kinase domain 3a8x 
Crystal Structure of PKCiota kinase domain 3ag9 
Complex of PKA with the bisubstrate protein kinase inhibitor ARC-1012 3agl 
Complex of PKA with the bisubstrate protein kinase inhibitor ARC-1039 3agm 
Complex of PKA with the bisubstrate protein kinase inhibitor ARC-670 3ama 
Protein kinase A sixfold mutant model of Aurora B with inhibitor JNJ-7706621 3amb 
Protein kinase A sixfold mutant model of Aurora B with inhibitor VX-680 3bwj 
Complex of PKA with the bisubstrate protein kinase inhibitor lead compound Arc-1034 3c4w 
Crystal Structure of G protein coupled receptor kinase 1 bound to ATP and magnesium chloride at 2.7A 3c4x 
Crystal Structure of G protein coupled receptor kinase 1 bound to ATP and magnesium chloride at 2.9A 3c4y 
Crystal Structure of Apo form of G protein coupled receptor kinase 1 at 7.51A 3c4z 
Crystal structure of G protein coupled receptor kinase 1 bound to ADP and magnesium chloride at 1.84A 3c50 
Crystal Structure of G protein coupled receptor kinase 1 bound to ADP and magnesium chloride at 2.6A 3c51 
Crystal structure of G protein coupled receptor kinase 1 bound to ADP and magnesium chloride at 3.55A 3cik 
Human GRK2 in Complex with Gbetagamma subunits 3cqu 
Crystal Structure of Akt-1 complexed with substrate peptide and inhibitor 3cqw 
Crystal Structure of Akt-1 complexed with substrate peptide and inhibitor 3d0e 
Crystal structure of human Akt2 in complex with GSK690693 3d9v 
CRYSTAL STRUCTURE OF ROCK I BOUND TO H-1152P A DI-METHYLATED VARIANT OF FASUDIL 3dnd 
cAMP-dependent protein kinase PKA catalytic subunit with PKI-5-24 3dne 
cAMP-dependent protein kinase PKA catalytic subunit with PKI-5-24 3e87 
Crystal structures of the kinase domain of AKT2 in complex with ATP-competitive inhibitors 3e88 
Crystal structures of the kinase domain of AKT2 in complex with ATP-competitive inhibitors 3e8c 
Crystal structures of the kinase domain of PKA in complex with ATP-competitive inhibitors 3e8d 
Crystal structures of the kinase domain of AKT2 in complex with ATP-competitive inhibitors 3e8e 
Crystal structures of the kinase domain of PKA in complex with ATP-competitive inhibitors 3fhi 
Crystal structure of a complex between the catalytic and regulatory (RI{alpha}) subunits of PKA 3fjq 
Crystal structure of cAMP-dependent protein kinase catalytic subunit alpha in complex with peptide inhibitor PKI alpha (6-25) 3g51 
Structural diversity of the active conformation of the N-terminal kinase domain of p90 ribosomal S6 kinase 2 3hdm 
Crystal structure of serum and glucocorticoid-regulated kinase 1 in complex with compound 1 3hdn 
Crystal structure of serum and glucocorticoid-regulated kinase 1 in complex with compound 2 3idb 
Crystal structure of (108-268)RIIb:C holoenzyme of cAMP-dependent protein kinase 3idc 
Crystal structure of (102-265)RIIb:C holoenzyme of cAMP-dependent protein kinase 3iw4 
Crystal structure of PKC alpha in complex with NVP-AEB071 3j4q 
Pseudo-atomic model of the AKAP18-PKA complex in a bent conformation derived from electron microscopy 3j4r 
Pseudo-atomic model of the AKAP18-PKA Complex in a linear conformation derived from electron microscopy 3kkv 
Structure of PKA with a protein Kinase B-selective inhibitor. 3krw 
Human GRK2 in complex with Gbetgamma subunits and balanol (soak) 3krx 
Human GRK2 in complex with Gbetgamma subunits and balanol (co-crystal) 3l9l 
Crystal structure of pka with compound 36 3l9m 
Crystal structure of PKAB3 (pka triple mutant V123A, L173M, Q181K) with compound 18 3l9n 
crystal structure of PKAB3 (pka triple mutant V123A, L173M, Q181K) with compound 27 3mv5 
Crystal structure of Akt-1-inhibitor complexes 3mvh 
Crystal structure of Akt-1-inhibitor complexes 3mvj 
Human cyclic AMP-dependent protein kinase PKA inhibitor complex 3ncz 
X-Ray Co-structure of Rho-Associated Protein Kinase (ROCK1) with a potent 2H-isoquinolin-1-one inhibitor 3ndm 
Crystal structure of Rho-Associated Protein Kinase (ROCK1) with a potent isoquinolone derivative 3nx8 
human cAMP dependent protein kinase in complex with phenol 3nyn 
Crystal Structure of G Protein-Coupled Receptor Kinase 6 in Complex with Sangivamycin 3nyo 
Crystal Structure of G Protein-Coupled Receptor Kinase 6 in Complex with AMP 3o7l 
Crystal Structure of phospholamban (1-19):PKA C-subunit:AMP-PNP:Mg2+ complex 3o96 
Crystal Structure of Human AKT1 with an Allosteric Inhibitor 3ocb 
Akt1 kinase domain with pyrrolopyrimidine inhibitor 3oog 
human cAMP-dependent protein kinase in complex with a small fragment 3ovv 
Human cAMP-dependent protein kinase in complex with an inhibitor 3ow3 
Discovery of dihydrothieno- and dihydrofuropyrimidines as potent pan Akt inhibitors 3ow4 
Discovery of dihydrothieno- and dihydrofuropyrimidines as potent pan Akt inhibitors 3owp 
Human cAMP-dependent protein kinase in complex with an inhibitor 3oxt 
Human cAMP-dependent protein kinase in complex with an inhibitor 3p0m 
Human cAMP-dependent protein kinase in complex with an inhibitor 3pfq 
Crystal Structure and Allosteric Activation of Protein Kinase C beta II 3poo 
human cAMP-dependent protein kinase in complex with an inhibitor 3psc 
Bovine GRK2 in complex with Gbetagamma subunits 3pvb 
Crystal structure of (73-244)RIa:C holoenzyme of cAMP-dependent Protein kinase 3pvu 
Bovine GRK2 in complex with Gbetagamma subunits and a selective kinase inhibitor (CMPD101) 3pvw 
Bovine GRK2 in complex with Gbetagamma subunits and a selective kinase inhibitor (CMPD103A) 3qal 
Crystal Structure of Arg280Ala mutant of Catalytic subunit of cAMP-dependent Protein Kinase 3qam 
Crystal Structure of Glu208Ala mutant of catalytic subunit of cAMP-dependent protein kinase 3qc9 
Crystal structure of cross-linked bovine GRK1 T8C/N480C double mutant complexed with ADP and Mg 3qfv 
MRCK beta in complex with TPCA-1 3qkk 
Spirochromane Akt Inhibitors 3qkl 
Spirochromane Akt Inhibitors 3qkm 
Spirocyclic sulfonamides as AKT inhibitors 3t8o 
Rhodopsin kinase (GRK1) L166K mutant at 2.5A resolution 3tku 
MRCK beta in complex with fasudil 3tnp 
Structure and Allostery of the PKA RIIb Tetrameric Holoenzyme 3tnq 
Structure and Allostery of the PKA RIIb Tetrameric Holoenzyme 3tv7 
Human Rho-associated protein kinase 1 (ROCK 1) in COMPLEX WITH RKI1342 3twj 
Rho-associated protein kinase 1 (ROCK 1) IN COMPLEX WITH RKI1447 3txo 
PKC eta kinase in complex with a naphthyridine 3uzs 
Structure of the C13.28 RNA Aptamer Bound to the G Protein-Coupled Receptor Kinase 2-Heterotrimeric G Protein Beta 1 and Gamma 2 Subunit Complex 3uzt 
Structure of the C13.18 RNA Aptamer in Complex with G Protein-Coupled Receptor Kinase 2 3v5w 
Human G Protein-Coupled Receptor Kinase 2 in Complex with Soluble Gbetagamma Subunits and Paroxetine 3v8s 
Human RHO-ASSOCIATED PROTEIN KINASE 1 (ROCK 1) IN COMPLEX WITH INDAZOLE DERIVATIVE (COMPOUND 18) 3vqh 
Bromine SAD partially resolves multiple binding modes for PKA inhibitor H-89 3we4 
3WE4 3wf5 
3WF5 3wf6 
3WF6 3wf7 
3WF7 3wf8 
3WF8 3wf9 
3WF9 3x2u 
3X2U 3x2v 
3X2V 3x2w 
3X2W 3zh8 
A novel small molecule aPKC inhibitor 3zo1 
The Synthesis and Evaluation of Diazaspirocyclic Protein Kinase Inhibitors 3zo2 
The Synthesis and Evaluation of Diazaspirocyclic Protein Kinase Inhibitors 3zo3 
The Synthesis and Evaluation of Diazaspirocyclic Protein Kinase Inhibitors 3zo4 
The Synthesis and Evaluation of Diazaspirocyclic Protein Kinase Inhibitors 4ae6 
A Crystal Structure of the Sperm Specific Isoform of Protein Kinase A (PKA) Subunit CalphaS Reveals a Binding Pocket for Hydrophobic Moieties 4ae9 
A Crystal Structure of the Sperm Specific Isoform of Protein Kinase A (PKA) Subunit CalphaS Reveals a Binding Pocket for Hydrophobic Moieties 4aw2 
Crystal structure of CDC42 binding protein kinase alpha (MRCK alpha) 4axa 
Structure of PKA-PKB chimera complexed with (1S)-2-amino-1-(4- chlorophenyl)-1-(4-(1H-pyrazol-4-yl)phenyl)ethan-1-ol 4c33 
PKA-S6K1 Chimera Apo 4c34 
PKA-S6K1 Chimera with Staurosporine bound 4c35 
PKA-S6K1 Chimera with compound 1 (NU1085) bound 4c36 
PKA-S6K1 Chimera with compound 15e (CCT147581) bound 4c37 
PKA-S6K1 Chimera with compound 21a (CCT196539) bound 4c38 
PKA-S6K1 Chimera with compound 21e (CCT239066) bound 4crs 
Human Protein Kinase N2 (PKN2, PRKCL2) in complex with ATPgammaS 4dc2 
Structure of PKC in Complex with a Substrate Peptide from Par-3 4dfx 
Crystal structure of myristoylated K7C catalytic subunit of cAMP-dependent protein kinase in complex with SP20 and AMP-PNP 4dfy 
Crystal structure of R194A mutant of cAMP-dependent protein kinase with unphosphorylated activation loop 4dfz 
Crystal structure of myristoylated K7C catalytic subunit of cAMP-dependent protein kinase in complex with SP20 4dg0 
Crystal structure of myristoylated WT catalytic subunit of cAMP-dependent protein kinase in complex with SP20 and AMP-PNP 4dg2 
Crystal structure of myristoylated WT catalytic subunit of cAMP-dependent protein kinase in complex with SP20 4dg3 
Crystal structure of R336A mutant of cAMP-dependent protein kinase with unphosphorylated turn motif. 4dh1 
Low temperature X-ray structure of cAMP dependent Protein Kinase A catalytic subunit with low Mg2+, ATP and IP20 4dh3 
Low temperature X-ray structure of cAMP dependent Protein Kinase A catalytic subunit with high Mg2+, ATP and IP20 4dh5 
Room temperature X-ray structure of cAMP dependent Protein Kinase A catalytic subunit with high Mg2+, ADP, Phosphate, and IP20 4dh7 
Low temperature X-ray structure of cAMP dependent Protein Kinase A catalytic subunit with high Mg2+, AMP-PNP and IP20' 4dh8 
Room temperature X-ray structure of cAMP dependent Protein Kinase A catalytic subunit with high Mg2+, AMP-PNP and IP20 4din 
Novel Localization and Quaternary Structure of the PKA RI beta Holoenzyme 4ejn 
Crystal structure of autoinhibited form of AKT1 in complex with N-(4-(5-(3-acetamidophenyl)-2-(2-aminopyridin-3-yl)-3H-imidazo[4,5-b]pyridin-3-yl)benzyl)-3-fluorobenzamide 4ekk 
Akt1 with AMP-PNP 4ekl 
Akt1 with GDC0068 4gv1 
PKB alpha in complex with AZD5363 4hpt 
Crystal structure of the catalytic subunit of cAMP-dependent protein kinase displaying complete phosphoryl transfer of AMP-PNP onto a substrate peptide 4hpu 
Crystal structure of the catalytic subunit of cAMP-dependent protein kinase displaying partial phosphoryl transfer of AMP-PNP onto a substrate peptide 4iac 
X-RAY structure of cAMP dependent protein kinase A in compelx with HIGH MG2+ concentration, AMP-PCP AND pseudo-substrate peptide SP20 4iad 
Low temperature X-ray Structure OF cAMP dependent protein kinase A in compelx with high Mg2+ concentration, ADP and phosphorylated peptide pSP20 4iaf 
Room temperature X-ray Structure OF cAMP dependent protein kinase A in complex with high Mg2+ concentration, ADP and phosphorylated peptide pSP20 4iai 
X-ray Structure of cAMP dependent protein kinase A in complex with high Ca2+ concentration, ADP and phosphorylated peptide pSP20 4iak 
Low temperature X-ray structure of cAMP dependent protein kinase A in complex with high Sr2+ concentration, ADP and phosphorylated peptide pSP20 4iay 
Room temperature X-ray Structure of cAMP dependent protein kinase A in complex with high Sr2+ concentration, ADP and phosphorylated peptide pSP20 4iaz 
Structure of cAMP dependent protein kinase A in complex with high Ba2+ concentration, ADP and phosphorylated peptide pSP20 4ib0 
X-ray Structure of cAMP dependent protein kinase A in complex with high Na+ concentration, ADP and phosphorylated peptide pSP20 4ib1 
Structure of cAMP dependent protein kinase A in complex with high K+ concentration, ADP and phosphorylated peptide pSP20 4ib3 
Structure of cAMP dependent protein kinase A in complex with ADP, phosphorylated peptide pSP20, and no metal 4ie9 
Bovine PKA C-alpha in complex with 3-pyridylmethyl-5-methyl-1H-pyrazole-3-carboxylate 4ij9 
Bovine PKA C-alpha in complex with 2-[[5-(4-pyridyl)-1H-1,2,4-triazol-3-yl]sulfanyl]-1-(2-thiophenyl)ethanone 4l42 
Crystal structures of human p70S6K1-PIF 4l43 
Crystal structures of human p70S6K1-T389A (form I) 4l44 
Crystal structures of human p70S6K1-T389A (form II) 4l45 
Crystal structures of human p70S6K1-T389E 4l46 
Crystal structures of human p70S6K1-WT 4l6q 
ROCK2 in complex with benzoxaborole 4l9i 
Bovine G Protein Coupled Receptor Kinase 1 in Complex with Paroxetine 4lqp 
4LQP 4lqq 
4LQQ 4lqs 
4LQS 4mk0 
Crystal structure of G protein-coupled receptor kinase 2 in complex with a a rationally designed paroxetine derivative 4nts 
4NTS 4ntt 
4NTT 4nus 
Rsk2 N-terminal kinase in complex with LJH685 4nw5 
Rsk2 N-terminal kinase in complex with 2-amino-7-substituted benzoxazole compound 8 4nw6 
Rsk2 N-terminal kinase in complex with 2-amino-7-substituted benzoxazole compound 27 4o21 
4O21 4o22 
4O22 4otd 
4OTD 4otg 
4OTG 4oth 
4OTH 4oti 
4OTI 4pni 
4PNI 4pnk 
4PNK 4q9z 
4Q9Z 4ra4 
4RA4 4ra5 
4RA5 4tnb 
4TNB 4tnd 
4TND 4uak 
4UAK 4ual 
4UAL 4uj1 
4UJ1 4uj2 
4UJ2 4uj9 
4UJ9 4uja 
4UJA 4ujb 
4UJB 4w7p 
4W7P 4wb5 
4WB5 4wb6 
4WB6 4wb7 
4WB7 4wb8 
4WB8 4wbb 
4WBB 4wbo 
4WBO 4wih 
4WIH 4wnk 
4WNK 4wot 
4WOT 4x6q 
4X6Q 4x6r 
4X6R 4xw4 
4XW4 4xw5 
4XW5 4xw6 
4XW6 4yhj 
4YHJ 4yvc 
4YVC 4yve 
4YVE 4yxr 
4YXR 4yxs 
4YXS 4z83 
4Z83 4z84 
4Z84 5bml 
5BML 5bx6 
5BX6 5bx7 
5BX7 5d9k 
5D9K 5d9l 
5D9L 5dyk 
5DYK 5f9e 
5F9E 5he0 
5HE0 5he1 
5HE1 5he2 
5HE2 5he3 
5HE3 5izf 
5IZF 5izj 
5IZJ 5j5x 
5J5X 5kcv 
5KCV 5li1 
5LI1 5li9 
5LI9 5lih 
5LIH 5loh 
5LOH - Links (links to other resources describing this domain)
-
INTERPRO IPR000961

