The domain within your query sequence starts at position 103 and ends at position 263; the E-value for the ZnMc domain shown below is 5.78e-60.
All catalytic sites are present in this domain. Check the literature (PubMed 96311273 ) for details.
NSPKWHSRIVTYRIVSYTSDLPRIVVDQIVKKALRMWSMQIPLNFKRVSWGTADIIIGFA RRDHGDSFPFDGPGNTLGHAFAPGPGLGGDAHFDKDEYWTDGEDAGVNFLFAATHEFGHS LGLSHSSVPGTVMYPTYQRDYSEDFSLTKDDIAGIQKLYGK
ZnMcZinc-dependent metalloprotease |
|---|
| SMART accession number: | SM00235 |
|---|---|
| Description: | Neutral zinc metallopeptidases. This alignment represents a subset of known subfamilies. Highest similarity occurs in the HExxH zinc-binding site/ active site. |
| Interpro abstract (IPR006026): | Over 70 metallopeptidase families have been identified to date. In these enzymes a divalent cation which is usually zinc, but may be cobalt, manganese or copper, activates the water molecule. The metal ion is held in place by amino acid ligands, usually three in number. In some families of co-catalytic metallopeptidases, two metal ions are observed in crystal structures ligated by five amino acids, with one amino acid ligating both metal ions. The known metal ligands are His, Glu, Asp or Lys. At least one other residue is required for catalysis, which may play an electrophillic role. Many metalloproteases contain an HEXXH motif, which has been shown in crystallographic studies to form part of the metal-binding site [ (PUBMED:7674922) ]. The HEXXH motif is relatively common, but can be more stringently defined for metalloproteases as 'abXHEbbHbc', where 'a' is most often valine or threonine and forms part of the S1' subsite in thermolysin and neprilysin, 'b' is an uncharged residue, and 'c' a hydrophobic residue. Proline is never found in this site, possibly because it would break the helical structure adopted by this motif in metalloproteases [ (PUBMED:7674922) ]. The majority of zinc-dependent metallopeptidases (with the notable exception of the carboxypeptidases) share a common pattern of primary structure [ (PUBMED:2914602) (PUBMED:1894005) ] in the part of their sequence involved in the binding of zinc, and can be grouped together as a superfamily,known as the metzincins, on the basis of this sequence similarity. They can be classified into around 40 distinct families [ (PUBMED:7674922) ]. This signature defines the metallopeptidases associated with MEROPS peptidase families: M7, M8, M10 (subfamilies A, B and C) and M12 (subfamily A) all of which are members of clan MA(M). |
| GO process: | proteolysis (GO:0006508) |
| GO function: | zinc ion binding (GO:0008270), metallopeptidase activity (GO:0008237) |
| Family alignment: |
There are 34425 ZnMc domains in 34168 proteins in SMART's nrdb database.
Click on the following links for more information.
- Evolution (species in which this domain is found)
-
Taxonomic distribution of proteins containing ZnMc domain.
This tree includes only several representative species. The complete taxonomic breakdown of all proteins with ZnMc domain is also avaliable.
Click on the protein counts, or double click on taxonomic names to display all proteins containing ZnMc domain in the selected taxonomic class.
- Cellular role (predicted cellular role)
-
Binding / catalysis: Peptidase; zinc-binding
- Literature (relevant references for this domain)
-
Primary literature is listed below; Automatically-derived, secondary literature is also avaliable.
- Gooley PR et al.
- The NMR structure of the inhibited catalytic domain of human stromelysin-1.
- Nat Struct Biol. 1994; 1: 111-8
- Display abstract
The three-dimensional structure of the catalytic domain of stromelysin-1 complexed with an N-carboxyl alkyl inhibitor has been determined by NMR methods. The global fold consists of three helices, a five stranded beta-sheet and a methionine located in a turn near the catalytic histidines, classifying stromelysin-1 as a metzincin. Stromelysin-1 is unique in having two independent zinc binding sites: a catalytic site and a structural site. The inhibitor binds in an extended conformation. The S1' subsite is a deep hydrophobic pocket, whereas S2' appears shallow and S3' open.
- Metabolism (metabolic pathways involving proteins which contain this domain)
-
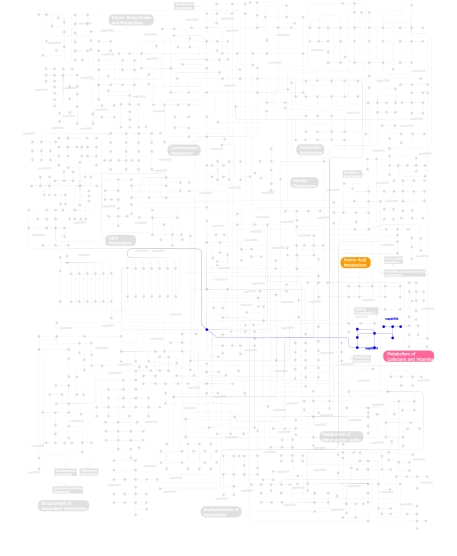
Click the image to view the interactive version of the map in iPath% proteins involved KEGG pathway ID Description 31.46 map05219 Bladder cancer 22.47 map04670 Leukocyte transendothelial migration 21.35 map04912 GnRH signaling pathway 8.99 map04310 Wnt signaling pathway 8.99 map03320 PPAR signaling pathway 3.37  map00780
map00780Biotin metabolism 3.37  map00310
map00310Lysine degradation This information is based on mapping of SMART genomic protein database to KEGG orthologous groups. Percentage points are related to the number of proteins with ZnMc domain which could be assigned to a KEGG orthologous group, and not all proteins containing ZnMc domain. Please note that proteins can be included in multiple pathways, ie. the numbers above will not always add up to 100%.
- Structure (3D structures containing this domain)
3D Structures of ZnMc domains in PDB
PDB code Main view Title 1a85 
MMP8 WITH MALONIC AND ASPARAGINE BASED INHIBITOR 1a86 
MMP8 WITH MALONIC AND ASPARTIC ACID BASED INHIBITOR 1af0 
SERRATIA PROTEASE IN COMPLEX WITH INHIBITOR 1akl 
ALKALINE PROTEASE FROM PSEUDOMONAS AERUGINOSA IFO3080 1ast 
STRUCTURE OF ASTACIN AND IMPLICATIONS FOR ACTIVATION OF ASTACINS AND ZINC-LIGATION OF COLLAGENASES 1ayk 
INHIBITOR-FREE CATALYTIC FRAGMENT OF HUMAN FIBROBLAST COLLAGENASE, NMR, 30 STRUCTURES 1b3d 
STROMELYSIN-1 1b8y 
X-RAY STRUCTURE OF HUMAN STROMELYSIN CATALYTIC DOMAIN COMPLEXED WITH NON-PEPTIDE INHIBITORS: IMPLICATIONS FOR INHIBITOR SELECTIVITY 1biw 
DESIGN AND SYNTHESIS OF CONFORMATIONALLY-CONSTRAINED MMP INHIBITORS 1bm6 
SOLUTION STRUCTURE OF THE CATALYTIC DOMAIN OF HUMAN STROMELYSIN-1 COMPLEXED TO A POTENT NON-PEPTIDIC INHIBITOR, NMR, 20 STRUCTURES 1bqo 
DISCOVERY OF POTENT, ACHIRAL MATRIX METALLOPROTEINASE INHIBITORS 1bqq 
CRYSTAL STRUCTURE OF THE MT1-MMP--TIMP-2 COMPLEX 1buv 
CRYSTAL STRUCTURE OF THE MT1-MMP-TIMP-2 COMPLEX 1bzs 
CRYSTAL STRUCTURE OF MMP8 COMPLEXED WITH HMR2909 1c3i 
HUMAN STROMELYSIN-1 CATALYTIC DOMAIN COMPLEXED WITH RO-26-2812 1c8t 
HUMAN STROMELYSIN-1 (E202Q) CATALYTIC DOMAIN COMPLEXED WITH RO-26-2812 1caq 
X-RAY STRUCTURE OF HUMAN STROMELYSIN CATALYTIC DOMAIN COMPLEXES WITH NON-PEPTIDE INHIBITORS: IMPLICATION FOR INHIBITOR SELECTIVITY 1cge 
CRYSTAL STRUCTURES OF RECOMBINANT 19-KDA HUMAN FIBROBLAST COLLAGENASE COMPLEXED TO ITSELF 1cgf 
CRYSTAL STRUCTURES OF RECOMBINANT 19-KDA HUMAN FIBROBLAST COLLAGENASE COMPLEXED TO ITSELF 1cgl 
STRUCTURE OF THE CATALYTIC DOMAIN OF FIBROBLAST COLLAGENASE COMPLEXED WITH AN INHIBITOR 1ciz 
X-RAY STRUCTURE OF HUMAN STROMELYSIN CATALYTIC DOMAIN COMPLEXES WITH NON-PEPTIDE INHIBITORS: IMPLICATION FOR INHIBITOR SELECTIVITY 1ck7 
GELATINASE A (FULL-LENGTH) 1cqr 
CRYSTAL STRUCTURE OF THE STROMELYSIN CATALYTIC DOMAIN AT 2.0 A RESOLUTION 1cxv 
STRUCTURE OF RECOMBINANT MOUSE COLLAGENASE-3 (MMP-13) 1d5j 
CRYSTAL STRUCTURE OF MMP3 COMPLEXED WITH A THIAZEPINE BASED INHIBITOR. 1d7x 
CRYSTAL STRUCTURE OF MMP3 COMPLEXED WITH A MODIFIED PROLINE SCAFFOLD BASED INHIBITOR. 1d8f 
CRYSTAL STRUCTURE OF MMP3 COMPLEXED WITH A PIPERAZINE BASED INHIBITOR. 1d8m 
CRYSTAL STRUCTURE OF MMP3 COMPLEXED WITH A HETEROCYCLE-BASED INHIBITOR 1eak 
Catalytic domain of proMMP-2 E404Q mutant 1eub 
SOLUTION STRUCTURE OF THE CATALYTIC DOMAIN OF HUMAN COLLAGENASE-3 (MMP-13) COMPLEXED TO A POTENT NON-PEPTIDIC SULFONAMIDE INHIBITOR 1fbl 
STRUCTURE OF FULL-LENGTH PORCINE SYNOVIAL COLLAGENASE (MMP1) REVEALS A C-TERMINAL DOMAIN CONTAINING A CALCIUM-LINKED, FOUR-BLADED BETA-PROPELLER 1fls 
SOLUTION STRUCTURE OF THE CATALYTIC FRAGMENT OF HUMAN COLLAGENASE-3 (MMP-13) COMPLEXED WITH A HYDROXAMIC ACID INHIBITOR 1fm1 
SOLUTION STRUCTURE OF THE CATALYTIC FRAGMENT OF HUMAN COLLAGENASE-3 (MMP-13) COMPLEXED WITH A HYDROXAMIC ACID INHIBITOR 1g05 
HETEROCYCLE-BASED MMP INHIBITOR WITH P2'SUBSTITUENTS 1g49 
A CARBOXYLIC ACID BASED INHIBITOR IN COMPLEX WITH MMP3 1g4k 
X-ray Structure of a Novel Matrix Metalloproteinase Inhibitor Complexed to Stromelysin 1g9k 
CRYSTAL STRUCTURE OF A PSYCHROPHILIC ALKALINE PROTEASE FROM PSEUDOMONAS TAC II 18 1gkc 
MMP9-inhibitor complex 1gkd 
MMP9 active site mutant-inhibitor complex 1go7 
The metzincin's methionine: PrtC M226C-E189K double mutant 1go8 
The metzincin's methionine: PrtC M226L mutant 1gxd 
proMMP-2/TIMP-2 complex 1h71 
Psychrophilic Protease from Pseudoalteromonas haloplanctis 1hfc 
1.56 ANGSTROM STRUCTURE OF MATURE TRUNCATED HUMAN FIBROBLAST COLLAGENASE 1hfs 
CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF HUMAN FIBROBLAST STROMELYSIN-1 INHIBITED WITH THE N-CARBOXY-ALKYL INHIBITOR L-764,004 1hov 
SOLUTION STRUCTURE OF A CATALYTIC DOMAIN OF MMP-2 COMPLEXED WITH SC-74020 1hv5 
CRYSTAL STRUCTURE OF THE STROMELYSIN-3 (MMP-11) CATALYTIC DOMAIN COMPLEXED WITH A PHOSPHINIC INHIBITOR 1hy7 
A CARBOXYLIC ACID BASED INHIBITOR IN COMPLEX WITH MMP3 1i73 
COMPLEX OF PRO-LEU-L-TRP PHOSPHONATE WITH THE CATALITIC DOMAIN OF MATRIX METALLO PROTEINASE-8 (MET80 FORM) 1i76 
COMPLEX OF 2-(BIPHENYL-4-SULFONYL)-1,2,3,4-TETRAHYDRO-ISOQUINOLINE-3-CARBOXYLIC ACID (D-TIC DERIVATIVE) WITH T CATALITIC DOMAIN OF MATRIX METALLO PROTEINASE-8 (MET80 FORM) 1iaa 
CRYSTAL STRUCTURES, SPECTROSCOPIC FEATURES, AND CATALYTIC PROPERTIES OF COBALT(II), COPPER(II), NICKEL(II), AND MERCURY(II) DERIVATIVES OF THE ZINC ENDOPEPTIDASE ASTACIN. A CORRELATION OF STRUCTURE AND PROTEOLYTIC ACTIVITY 1iab 
CRYSTAL STRUCTURES, SPECTROSCOPIC FEATURES, AND CATALYTIC PROPERTIES OF COBALT(II), COPPER(II), NICKEL(II), AND MERCURY(II) DERIVATIVES OF THE ZINC ENDOPEPTIDASE ASTACIN. A CORRELATION OF STRUCTURE AND PROTEOLYTIC ACTIVITY 1iac 
REFINED 1.8 ANGSTROMS X-RAY CRYSTAL STRUCTURE OF ASTACIN, A ZINC-ENDOPEPTIDASE FROM THE CRAYFISH ASTACUS ASTACUS L. STRUCTURE DETERMINATION, REFINEMENT, MOLECULAR STRUCTURE AND COMPARISON WITH THERMOLYSIN 1iad 
REFINED 1.8 ANGSTROMS X-RAY CRYSTAL STRUCTURE OF ASTACIN, A ZINC-ENDOPEPTIDASE FROM THE CRAYFISH ASTACUS ASTACUS L. STRUCTURE DETERMINATION, REFINEMENT, MOLECULAR STRUCTURE AND COMPARISON TO THERMOLYSIN 1iae 
CRYSTAL STRUCTURES, SPECTROSCOPIC FEATURES, AND CATALYTIC PROPERTIES OF COBALT(II), COPPER(II), NICKEL(II), AND MERCURY(II) DERIVATIVES OF THE ZINC ENDOPEPTIDASE ASTACIN. A CORRELATION OF STRUCTURE AND PROTEOLYTIC ACTIVITY 1jan 
COMPLEX OF PRO-LEU-GLY-HYDROXYLAMINE WITH THE CATALYTIC DOMAIN OF MATRIX METALLO PROTEINASE-8 (PHE79 FORM) 1jao 
COMPLEX OF 3-MERCAPTO-2-BENZYLPROPANOYL-ALA-GLY-NH2 WITH THE CATALYTIC DOMAIN OF MATRIX METALLO PROTEINASE-8 (MET80 FORM) 1jap 
COMPLEX OF PRO-LEU-GLY-HYDROXYLAMINE WITH THE CATALYTIC DOMAIN OF MATRIX METALLO PROTEINASE-8 (MET80 FORM) 1jaq 
COMPLEX OF 1-HYDROXYLAMINE-2-ISOBUTYLMALONYL-ALA-GLY-NH2 WITH THE CATALYTIC DOMAIN OF MATRIX METALLO PROTEINASE-8 (MET80 FORM) 1jh1 
Crystal Structure of MMP-8 complexed with a 6H-1,3,4-thiadiazine derived inhibitor 1jiw 
Crystal structure of the APR-APRin complex 1jiz 
Crystal Structure Analysis of human Macrophage Elastase MMP-12 1jj9 
Crystal Structure of MMP8-Barbiturate Complex Reveals Mechanism for Collagen Substrate Recognition 1jk3 
Crystal structure of human MMP-12 (Macrophage Elastase) at true atomic resolution 1k7g 
PrtC from Erwinia chrysanthemi 1k7i 
PrtC from Erwinia chrysanthemi: Y228F mutant 1k7q 
PrtC from Erwinia chrysanthemi: E189A mutant 1kap 
THREE-DIMENSIONAL STRUCTURE OF THE ALKALINE PROTEASE OF PSEUDOMONAS AERUGINOSA: A TWO-DOMAIN PROTEIN WITH A CALCIUM BINDING PARALLEL BETA ROLL MOTIF 1kbc 
PROCARBOXYPEPTIDASE TERNARY COMPLEX 1l6j 
Crystal structure of human matrix metalloproteinase MMP9 (gelatinase B). 1mmb 
COMPLEX OF BB94 WITH THE CATALYTIC DOMAIN OF MATRIX METALLOPROTEINASE-8 1mmp 
MATRILYSIN COMPLEXED WITH CARBOXYLATE INHIBITOR 1mmq 
MATRILYSIN COMPLEXED WITH HYDROXAMATE INHIBITOR 1mmr 
MATRILYSIN COMPLEXED WITH SULFODIIMINE INHIBITOR 1mnc 
STRUCTURE OF HUMAN NEUTROPHIL COLLAGENASE REVEALS LARGE S1' SPECIFICITY POCKET 1o0q 
Crystal structure of a cold adapted alkaline protease from Pseudomonas TAC II 18, co-crystallized with 1 mM EDTA 1o0t 
CRYSTAL STRUCTURE OF A COLD ADAPTED ALKALINE PROTEASE FROM PSEUDOMONAS TAC II 18, CO-CRYSTALLIZED WITH 5 mM EDTA (5 DAYS) 1om6 
CRYSTAL STRUCTURE OF A COLD ADAPTED ALKALINE PROTEASE FROM PSEUDOMONAS TAC II 18, CO-CRYSTALLIZED WITH 5mM EDTA (2 MONTHS) 1om7 
CRYSTAL STRUCTURE OF A COLD ADAPTED ALKALINE PROTEASE FROM PSEUDOMONAS TAC II 18, SOAKED IN 85 mM EDTA 1om8 
CRYSTAL STRUCTURE OF A COLD ADAPTED ALKALINE PROTEASE FROM PSEUDOMONAS TAC II 18, CO-CRYSTALLYZED WITH 10 mM EDTA 1omj 
CRYSTAL STRUCTURE OF A PSYCHROPHILIC ALKALINE PROTEASE FROM PSEUDOMONAS TAC II 18 1oo9 
Orientation in Solution of MMP-3 Catalytic Domain and N-TIMP-1 from Residual Dipolar Couplings 1os2 
Ternary enzyme-product-inhibitor complexes of human MMP12 1os9 
Binary enzyme-product complexes of human MMP12 1q3a 
Crystal structure of the catalytic domain of human matrix metalloproteinase 10 1qia 
CRYSTAL STRUCTURE OF STROMELYSIN CATALYTIC DOMAIN 1qib 
CRYSTAL STRUCTURE OF GELATINASE A CATALYTIC DOMAIN 1qic 
CRYSTAL STRUCTURE OF STROMELYSIN CATALYTIC DOMAIN 1qji 
Structure of astacin with a transition-state analogue inhibitor 1qjj 
Structure of astacin with a hydroxamic acid inhibitor 1rm8 
Crystal structure of the catalytic domain of MMP-16/MT3-MMP: Characterization of MT-MMP specific features 1rmz 
Crystal structure of the catalytic domain of human MMP12 complexed with the inhibitor NNGH at 1.3 A resolution 1ros 
Crystal structure of MMP-12 complexed to 2-(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)ethyl-4-(4-ethoxy[1,1-biphenyl]-4-yl)-4-oxobutanoic acid 1sat 
CRYSTAL STRUCTURE OF THE 50 KDA METALLO PROTEASE FROM S. MARCESCENS 1slm 
CRYSTAL STRUCTURE OF FIBROBLAST STROMELYSIN-1: THE C-TRUNCATED HUMAN PROENZYME 1sln 
CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF HUMAN FIBROBLAST STROMELYSIN-1 INHIBITED WITH THE N-CARBOXY-ALKYL INHIBITOR L-702,842 1smp 
CRYSTAL STRUCTURE OF A COMPLEX BETWEEN SERRATIA MARCESCENS METALLO-PROTEASE AND AN INHIBITOR FROM ERWINIA CHRYSANTHEMI 1srp 
STRUCTURAL ANALYSIS OF SERRATIA PROTEASE 1su3 
X-ray structure of human proMMP-1: New insights into collagenase action 1uea 
MMP-3/TIMP-1 COMPLEX 1ums 
STROMELYSIN-1 CATALYTIC DOMAIN WITH HYDROPHOBIC INHIBITOR BOUND, PH 7.0, 32OC, 20 MM CACL2, 15% ACETONITRILE; NMR ENSEMBLE OF 20 STRUCTURES 1umt 
STROMELYSIN-1 CATALYTIC DOMAIN WITH HYDROPHOBIC INHIBITOR BOUND, PH 7.0, 32OC, 20 MM CACL2, 15% ACETONITRILE; NMR AVERAGE OF 20 STRUCTURES MINIMIZED WITH RESTRAINTS 1usn 
CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF HUMAN FIBROBLAST STROMELYSIN-1 INHIBITED WITH THIADIAZOLE INHIBITOR PNU-142372 1utt 
Crystal Structure of MMP-12 complexed to 2-(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)ethyl-4-(4-ethoxy[1,1-biphenyl]-4-yl)-4-oxobutanoic acid 1utz 
Crystal Structure of MMP-12 complexed to (2R)-3-({[4-[(pyridin-4-yl)phenyl]-thien-2-yl}carboxamido)(phenyl)propanoic acid 1xuc 
Matrix metalloproteinase-13 complexed with non-zinc binding inhibitor 1xud 
Matrix metalloproteinase-13 complexed with non-zinc binding inhibitor 1xur 
Matrix metalloproteinase-13 complexed with non-zinc binding inhibitor 1y93 
Crystal structure of the catalytic domain of human MMP12 complexed with acetohydroxamic acid at atomic resolution 1ycm 
Solution Structure of matrix metalloproteinase 12 (MMP12) in the presence of N-Isobutyl-N-[4-methoxyphenylsulfonyl]glycyl hydroxamic acid (NNGH) 1you 
Crystal structure of the catalytic domain of MMP-13 complexed with a potent pyrimidinetrione inhibitor 1z3j 
Solution Structure of MMP12 in the presence of N-isobutyl-N-4-methoxyphenylsulfonyl]glycyl hydroxamic acid (NNGH) 1zp5 
Crystal structure of the complex between MMP-8 and a N-hydroxyurea inhibitor 1zs0 
Crystal structure of the complex between MMP-8 and a phosphonate inhibitor (S-enantiomer) 1ztq 
Crystal structure of the catalytic domain of MMP-13 complexed with WAY-033 1zvx 
Crystal structure of the complex between MMP-8 and a phosphonate inhibitor (R-enantiomer) 2ayk 
INHIBITOR-FREE CATALYTIC FRAGMENT OF HUMAN FIBROBLAST COLLAGENASE, NMR, MINIMIZED AVERAGE STRUCTURE 2clt 
Crystal structure of the active form (full-length) of human fibroblast collagenase. 2d1n 
Collagenase-3 (MMP-13) complexed to a hydroxamic acid inhibitor 2d1o 
Stromelysin-1 (MMP-3) complexed to a hydroxamic acid inhibitor 2ddy 
Solution Structure of Matrilysin (MMP-7) Complexed to Constraint Conformational Sulfonamide Inhibitor 2e2d 
Flexibility and variability of TIMP binding: X-ray structure of the complex between collagenase-3/MMP-13 and TIMP-2 2hu6 
Crystal structure of human MMP-12 in complex with acetohydroxamic acid and a bicyclic inhibitor 2j0t 
Crystal Structure of the Catalytic Domain of MMP-1 in Complex with the Inhibitory Domain of TIMP-1 2jnp 
Solution structure of matrix metalloproteinase 3 (MMP-3) in the presence of N-isobutyl-N-[4-methoxyphenylsulfonyl]glycyl hydroxamic acid (NNGH) 2jsd 
Solution structure of MMP20 complexed with NNGH 2jt5 
solution structure of matrix metalloproteinase 3 (MMP-3) in the presence of n-hydroxy-2-[n-(2-hydroxyethyl)biphenyl-4-sulfonamide] hydroxamic acid (MLC88) 2jt6 
Solution structure of matrix metalloproteinase 3 (MMP-3) in the presence of 3-4'-cyanobyphenyl-4-yloxy)-n-hdydroxypropionamide (MMP-3 inhibitor VII) 2k2g 
Solution structure of the wild-type catalytic domain of human matrix metalloproteinase 12 (MMP-12) in complex with a tight-binding inhibitor 2k9c 
Paramagnetic shifts in solid-state NMR of Proteins to elicit structural information 2krj 
High-Resolution Solid-State NMR Structure of a 17.6 kDa Protein 2mlr 
2MLR 2mls 
2MLS 2mze 
2MZE 2mzh 
2MZH 2mzi 
2MZI 2n8r 
2N8R 2ovx 
MMP-9 active site mutant with barbiturate inhibitor 2ovz 
MMP-9 active site mutant with phosphinate inhibitor 2ow0 
MMP-9 active site mutant with iodine-labeled carboxylate inhibitor 2ow1 
MMP-9 active site mutant with trifluoromethyl hydroxamate inhibitor 2ow2 
MMP-9 active site mutant with difluoro butanoic acid inhibitor 2ow9 
Crystal structure analysis of the MMP13 catalytic domain in complex with specific inhibitor 2oxu 
Uninhibited form of human MMP-12 2oxw 
Human MMP-12 complexed with the peptide IAG 2oxz 
Human MMP-12 in complex with two peptides PQG and IAG 2oy2 
Human MMP-8 in complex with peptide IAG 2oy4 
Uninhibited human MMP-8 2ozr 
MMP13 Catalytic Domain Complexed with 4-{[1-methyl-2,4-dioxo-6-(3-phenylprop-1-yn-1-yl)-1,4-dihydroquinazolin-3(2H)-yl]methyl}benzoic acid 2pjt 
Crystal structure of the catalytic domain of MMP-13 complexed with WAY-344 2poj 
NMR Solution Structure of the Inhibitor-Free State of Macrophage Metalloelastase (MMP-12) 2srt 
CATALYTIC DOMAIN OF HUMAN STROMELYSIN-1 AT PH 5.5 AND 40OC COMPLEXED WITH INHIBITOR 2tcl 
STRUCTURE OF THE CATALYTIC DOMAIN OF HUMAN FIBROBLAST COLLAGENASE COMPLEXED WITH AN INHIBITOR 2usn 
CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF HUMAN FIBROBLAST STROMELYSIN-1 INHIBITED WITH THIADIAZOLE INHIBITOR PNU-141803 2w0d 
Does a Fast Nuclear Magnetic Resonance Spectroscopy- and X-Ray Crystallography Hybrid Approach Provide Reliable Structural Information of Ligand-Protein Complexes? A Case Study of Metalloproteinases. 2wo8 
MMP12 complex with a beta hydroxy carboxylic acid 2wo9 
MMP12 complex with a beta hydroxy carboxylic acid 2woa 
MMP12 complex with a beta hydroxy carboxylic acid 2xs3 
Structure of karilysin catalytic MMP domain 2xs4 
Structure of karilysin catalytic MMP domain in complex with magnesium 2y6c 
The Discovery of MMP7 inhibitors Exploiting a Novel Selectivity Trigger 2y6d 
The Discovery of MMP7 Inhibitors Exploiting a Novel Selectivity Trigger 2yig 
MMP13 in complex with a novel selective non zinc binding inhibitor 2z2d 
Solution structure of human macrophage elastase (MMP-12) catalytic domain complexed with a gamma-keto butanoic acid inhibitor 3ayk 
CATALYTIC FRAGMENT OF HUMAN FIBROBLAST COLLAGENASE COMPLEXED WITH CGS-27023A, NMR, MINIMIZED AVERAGE STRUCTURE 3ayu 
Crystal structure of MMP-2 active site mutant in complex with APP-drived decapeptide inhibitor 3ba0 
Crystal structure of full-length human MMP-12 3dng 
Crystal structure of the complex between MMP-8 and a non-zinc chelating inhibitor 3dpe 
Crystal structure of the complex between MMP-8 and a non-zinc chelating inhibitor 3dpf 
Crystal structure of the complex between MMP-8 and a non-zinc chelating inhibitor 3edg 
Crystal structure of bone morphogenetic protein 1 protease domain 3edh 
Crystal structure of bone morphogenetic protein 1 protease domain in complex with partially bound DMSO 3edi 
Crystal structure of tolloid-like protease 1 (TLL-1) protease domain 3ehx 
Crystal structure of the catalytic domain of human MMP12 complexed with the inhibitor (R)-2-(biphenyl-4-ylsulfonamido)-4-methylpentanoic acid 3ehy 
Crystal structure of the catalytic domain of human MMP12 complexed with the inhibitor (R)-2-(4-methoxyphenylsulfonamido)propanoic acid 3elm 
Crystal Structure of MMP-13 Complexed with Inhibitor 24f 3f15 
Crystal structure of the catalytic domain of human mmp12 complexed with the inhibitor (S)-N-(2,3-dihydroxypropyl)-4-methoxy-N-(2-nitroso-2-oxoethyl)benzenesulfonamide 3f16 
Crystal structure of the catalytic domain of human MMP12 complexed with the inhibitor (R)-N-(3-hydroxy-1-nitroso-1-oxopropan-2-yl)-4-methoxybenzenesulfonamide 3f17 
Crystal structure of the catalytic domain of human MMP12 complexed with the inhibitor N-(2-nitroso-2-oxoethyl)biphenyl-4-sulfonamide 3f18 
Crystal structure of the catalytic domain of human MMP12 complexed with the inhibitor 4-fluoro-N-(2-hydroxyethyl)-N-(2-nitroso-2-oxoethyl)benzenesulfonamide 3f19 
Crystal structure of the catalytic domain of human MMP12 complexed with the inhibitor 4-fluoro-N-(2-nitroso-2-oxoethyl)benzenesulfonamide 3f1a 
Crystal structure of the catalytic domain of human MMP12 complexed with the inhibitor N-(2-nitroso-2-oxoethyl)benzenesulfonamide 3hb2 
PrtC methionine mutants: M226I 3hbu 
PrtC methionine mutants: M226H DESY 3hbv 
PrtC methionine mutants: M226A in-house 3hda 
PrtC methionine mutants: M226A_DESY 3i7g 
MMP-13 in complex with a non zinc-chelating inhibitor 3i7i 
MMP-13 in complex with a non zinc-chelating inhibitor 3kec 
Crystal Structure of Human MMP-13 complexed with a phenyl-2H-tetrazole compound 3kej 
Crystal Structure of Human MMP-13 complexed with a (pyridin-4-yl)-2H-tetrazole compound 3kek 
Crystal Structure of Human MMP-13 complexed with a (pyridin-4-yl)-2H-tetrazole compound 3kry 
Crystal structure of MMP-13 in complex with SC-78080 3lik 
Human MMP12 in complex with non-zinc chelating inhibitor 3lil 
Human MMP12 in complex with non-zinc chelating inhibitor 3lir 
Human MMP12 in complex with non-zinc chelating inhibitor 3ljg 
Human MMP12 in complex with non-zinc chelating inhibitor 3ljz 
Crystal Structure of Human MMP-13 complexed with an Amino-2-indanol compound 3lk8 
Crystal structure of the catalytic domain of human MMP12 complexed with the inhibitor paramethoxy-sulfonyl-glycine hydroxamate 3lka 
Catalytic domain of human MMP-12 complexed with hydroxamic acid and paramethoxy-sulfonyl amide 3lq0 
Zymogen structure of crayfish astacin metallopeptidase 3lqb 
Crystal structure of the hatching enzyme ZHE1 from the zebrafish Danio rerio 3ma2 
Complex membrane type-1 matrix metalloproteinase (MT1-MMP) with tissue inhibitor of metalloproteinase-1 (TIMP-1) 3n2u 
Crystal structure of the catalytic domain of human MMP12 complexed with the inhibitor N-hydroxy-2-(4-methoxy-N(2-(3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yloxy)ethyl)phenylsulfonamido)acetamide 3n2v 
Crystal structure of the catalytic domain of human MMP12 complexed with the inhibitor N-hydroxy-2-(N-hydroxyethyl)biphenyl-4-ylsulfonamido)acetamide 3nx7 
Crystal structure of the catalytic domain of human MMP12 complexed with the inhibitor N-Hydroxy-2-(N-(2-hydroxyethyl)4-methoxyphenylsulfonamido)acetamide 3o2x 
MMP-13 in complex with selective tetrazole core inhibitor 3ohl 
catalytic domain of stromelysin-1 in complex with N-Hydroxy-2-(4-methoxy-N-(pyridine-3-ylmethyl)phenylsulfonamido)acetamide 3oho 
catalytic domain of stromelysin-1 in complex with N-Hydroxy-2-(4-methylphenylsulfonamido)acetamide 3p24 
Structure of profragilysin-3 from Bacteroides fragilis 3rts 
Human MMP-12 catalytic domain in complex with*N*-Hydroxy-2-(2-phenylethylsulfonamido)acetamide 3rtt 
Human MMP-12 catalytic domain in complex with*(R)-N*-Hydroxy-1-(phenethylsulfonyl)pyrrolidine-2-carboxamide 3shi 
Crystal structure of human MMP1 catalytic domain at 2.2 A resolution 3ts4 
Human MMP12 in complex with L-glutamate motif inhibitor 3tsk 
Human MMP12 in complex with L-glutamate motif inhibitor 3tt4 
Human MMP8 in complex with L-glutamate motif inhibitor 3tvc 
Human MMP13 in complex with L-glutamate motif inhibitor 3u1r 
Structure Analysis of A New Psychrophilic Marine Protease 3usn 
STRUCTURE OF THE CATALYTIC DOMAIN OF HUMAN FIBROBLAST STROMELYSIN-1 INHIBITED WITH THE THIADIAZOLE INHIBITOR IPNU-107859, NMR, 1 STRUCTURE 3uvc 
MMP12 in a complex with the dimeric adduct: 5-(5-phenylhydantoin)-5-phenylhydantoin 3v96 
Complex of matrix metalloproteinase-10 catalytic domain (MMP-10cd) with tissue inhibitor of metalloproteinases-1 (TIMP-1) 3vi1 
Crystal structure of Pseudomonas aerginosa alkaline protease complexed with Substance P(1-6) 3vtg 
High choriolytic enzyme 1 (HCE-1), a hatching enzyme zinc-protease from Oryzias latipes (Medaka fish) 3wv1 
3WV1 3wv2 
3WV2 3wv3 
3WV3 3zxh 
MMP-13 complexed with 2-Napthylsulfonamide hydroxamic acid inhibitor 456c 
CRYSTAL STRUCTURE OF COLLAGENASE-3 (MMP-13) COMPLEXED TO A DIPHENYL-ETHER SULPHONE BASED HYDROXAMIC ACID 4a7b 
MMP13 IN COMPLEX WITH A NOVEL SELECTIVE NON ZINC BINDING INHIBITOR CMPD22 4auo 
Crystal structure of MMP-1(E200A) in complex with a triple-helical collagen peptide 4ayk 
CATALYTIC FRAGMENT OF HUMAN FIBROBLAST COLLAGENASE COMPLEXED WITH CGS-27023A, NMR, 30 STRUCTURES 4dpe 
Structure of MMP3 complexed with a platinum-based inhibitor. 4efs 
Human MMP12 in complex with L-glutamate motif inhibitor 4fu4 
Human collagenase 3 (MMP-13) with peptide from pro-domain 4fvl 
Human collagenase 3 (MMP-13) full form with peptides from pro-domain 4g0d 
Human collagenase 3 (MMP-13) full form with peptides from pro-domain 4g9l 
Structure of MMP3 complexed with NNGH inhibitor. 4gql 
Crystal structure of the catalytic domain of Human MMP12 in complex with selective phosphinic inhibitor RXP470.1 4gr0 
Crystal structure of the catalytic domain of Human MMP12 in complex with selective phosphinic inhibitor RXP470B 4gr3 
Crystal structure of the catalytic domain of Human MMP12 in complex with selective phosphinic inhibitor RXP470A 4gr8 
Crystal structure of the catalytic domain of Human MMP12 in complex with selective phosphinic inhibitor RXP470C 4guy 
Human MMP12 catalytic domain in complex with*N*-Hydroxy-2-(2-(4-methoxyphenyl)ethylsulfonamido)acetamide 4gwm 
Crystal structure of human promeprin beta 4gwn 
Crystal structure of human mature meprin beta 4h1q 
Crystal structure of mutant MMP-9 catalytic domain in complex with a twin inhibitor. 4h2e 
Crystal structure of an MMP twin inhibitor complexing two MMP-9 catalytic domains 4h30 
Crystal structure of the catalytic domain of MMP-12 in complex with a twin inhibitor. 4h3x 
Crystal structure of an MMP broad spectrum hydroxamate based inhibitor CC27 in complex with the MMP-9 catalytic domain 4h49 
Crystal structure of the catalytic domain of MMP-12 in complex with a twin inhibitor. 4h76 
Crystal structure of the catalytic domain of Human MMP12 in complex with a broad spectrum hydroxamate inhibitor 4h82 
Crystal structure of mutant MMP-9 catalytic domain in complex with a twin inhibitor. 4h84 
Crystal structure of the catalytic domain of Human MMP12 in complex with a selective carboxylate based inhibitor. 4hma 
Crystal structure of an MMP twin carboxylate based inhibitor LC20 in complex with the MMP-9 catalytic domain 4i03 
Human MMP12 in complex with a PEG-linked bifunctional L-glutamate motif inhibitor 4i35 
The crystal structure of serralysin 4ijo 
Unraveling hidden allosteric regulatory sites in structurally homologues metalloproteases 4ilw 
Complex of matrix metalloproteinase-10 catalytic domain (MMP-10cd) with tissue inhibitor of metalloproteinases-2 (TIMP-2) 4in9 
Structure of karilysin MMP-like catalytic domain in complex with inhibitory tetrapeptide SWFP 4ja1 
Structure of MMP3 complexed with a platinum-based inhibitor 4jij 
Crystal structure of an inactive mutant of MMP-9 catalytic domain in complex with a fluorogenic synthetic peptidic substrate 4jp4 
Mmp13 in complex with a reverse hydroxamate Zn-binder 4jpa 
Mmp13 in complex with a piperazine hydantoin ligand 4jqg 
Crystal structure of an inactive mutant of MMP-9 catalytic domain in complex with a fluorogenic synthetic peptidic substrate with a fluorine atom. 4l19 
4L19 4on1 
Crystal Structure of metalloproteinase-II from Bacteroides fragilis 4qkz 
4QKZ 4r3v 
4R3V 4wzv 
4WZV 4xct 
4XCT 5bot 
5BOT 5boy 
5BOY 5bpa 
5BPA 5cuh 
5CUH 5cxa 
5CXA 5czm 
5CZM 5czw 
5CZW 5d2b 
5D2B 5d3c 
5D3C 5d7w 
5D7W 5h8x 
5H8X 5i0l 
5I0L 5i12 
5I12 5i2z 
5I2Z 5i3m 
5I3M 5i43 
5I43 5i4o 
5I4O 5l79 
5L79 5l7f 
5L7F 5lab 
5LAB 830c 
COLLAGENASE-3 (MMP-13) COMPLEXED TO A SULPHONE-BASED HYDROXAMIC ACID 966c 
CRYSTAL STRUCTURE OF FIBROBLAST COLLAGENASE-1 COMPLEXED TO A DIPHENYL-ETHER SULPHONE BASED HYDROXAMIC ACID - Links (links to other resources describing this domain)
-
INTERPRO IPR006026

