The domain within your query sequence starts at position 212 and ends at position 501; the E-value for the STYKc domain shown below is 1e-25.
ECVGKGRYGEVWRGSWQGENVAVKIFSSRDEKSWFRETELYNTVMLRHENILGFIASDMT SRHSSTQLWLITHYHEMGSLYDYLQLTTLDTVSCLRIVLSIASGLAHLHIEIFGTQGKSA IAHRDLKSKNILVKKNGQCCIADLGLAVMHSQSTNQLDVGNNPRVGTKRYMAPEVLDETI QVDCFDSYKRVDIWAFGLVLWEVARRMVSNGIVEDYKPPFYDVVPNDPSFEDMRKVVCVD QQRPNIPNRWFSDPTLTSLAKLMKECWYQNPSARLTALRIKKTLTKIDNS
The domain was found using the schnipsel database
STYKcProtein kinase; unclassified specificity. |
|---|
| SMART accession number: | SM00221 |
|---|---|
| Description: | Phosphotransferases. The specificity of this class of kinases can not be predicted. Possible dual-specificity Ser/Thr/Tyr kinase. |
| Family alignment: |
There are 7538 STYKc domains in 4685 proteins in SMART's nrdb database.
Click on the following links for more information.
- Evolution (species in which this domain is found)
-
Taxonomic distribution of proteins containing STYKc domain.
This tree includes only several representative species. The complete taxonomic breakdown of all proteins with STYKc domain is also avaliable.
Click on the protein counts, or double click on taxonomic names to display all proteins containing STYKc domain in the selected taxonomic class.
- Literature (relevant references for this domain)
-
Primary literature is listed below; Automatically-derived, secondary literature is also avaliable.
- Engh RA, Girod A, Kinzel V, Huber R, Bossemeyer D
- Crystal structures of catalytic subunit of cAMP-dependent protein kinase in complex with isoquinolinesulfonyl protein kinase inhibitors H7, H8, and H89. Structural implications for selectivity.
- J Biol Chem. 1996; 271: 26157-64
- Display abstract
The discovery of several hundred different protein kinases involved in highly diverse cellular signaling pathways is in stark contrast to the much smaller number of known modulators of cell signaling. Of these, the H series protein kinase inhibitors (1-(5-isoquinolinesulfonyl)-2-methylpiperazine (H7), N-[2-(methylamino)ethyl]-5-isoquinolinesulfonamide (H8) N-[2-(p-Bromocinnamylamino)ethyl]-5-isoquinolinesulfonamide (H89)) are frequently used to block signaling pathways in studies of cellular regulation. To elucidate inhibition mechanisms at atomic resolution and to enable structure-based drug design of potential therapeutic modulators of signaling pathways, we determined the crystal structures of corresponding complexes with the cAPK catalytic subunit. Complexes with H7 and H8 (2.2 A) and with H89 (2.3 A) define the binding mode of the isoquinoline-sulfonamide derivatives in the ATP-binding site while demonstrating effects of ligand-induced structural change. Specific interactions between the enzyme and the inhibitors include the isoquinoline ring nitrogen ligating to backbone amide of Val-123 and an inhibitor side chain amide bonding to the backbone carbonyl of Glu-170. The conservation of the ATP-binding site of protein kinases allows evaluation of factors governing general selectivity of these inhibitors among kinases. These results should assist efforts in the design of protein kinase inhibitors with specific properties.
- Mohammadi M, Schlessinger J, Hubbard SR
- Structure of the FGF receptor tyrosine kinase domain reveals a novel autoinhibitory mechanism.
- Cell. 1996; 86: 577-87
- Display abstract
The crystal structure of the tyrosine kinase domain of fibroblast growth factor receptor 1 (FGFR1K) has been determined in its unliganded form to 2.0 angstroms resolution and in complex with with an ATP analog to 2.3 angstrosms A resolution. Several features distinguish the structure of FGFR1K from that of the tyrosine kinase domain of the insulin receptor. Residues in the activation loop of FGFR1K appear to interfere with substrate peptide binding but not with ATP binding, revealing a second and perhaps more general autoinhibitory mechanism for receptor tyrosine kinases. In addition, a dimeric form of FGFR1K observed in the crystal structure may provide insights into the molecular mechanisms by which FGF receptors are activated. Finally, the structure provides a basis for rationalizing the effects of kinase mutations in FGF receptors that lead to developmental disorders in nematodes and humans.
- Hanks SK, Hunter T
- Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification.
- FASEB J. 1995; 9: 576-96
- Display abstract
The eukaryotic protein kinases make up a large superfamily of homologous proteins. They are related by virtue of their kinase domains (also known as catalytic domains), which consist of approximately 250-300 amino acid residues. The kinase domains that define this group of enzymes contain 12 conserved subdomains that fold into a common catalytic core structure, as revealed by the 3-dimensional structures of several protein-serine kinases. There are two main subdivisions within the superfamily: the protein-serine/threonine kinases and the protein-tyrosine kinases. A classification scheme can be founded on a kinase domain phylogeny, which reveals families of enzymes that have related substrate specificities and modes of regulation.
- Owen DJ, Noble ME, Garman EF, Papageorgiou AC, Johnson LN
- Two structures of the catalytic domain of phosphorylase kinase: an active protein kinase complexed with substrate analogue and product.
- Structure. 1995; 3: 467-82
- Display abstract
BACKGROUND: Control of intracellular events by protein phosphorylation is promoted by specific protein kinases. All the known protein kinase possess a common structure that defines a catalytically competent entity termed the 'kinase catalytic core'. Within this common structural framework each kinase displays its own unique substrate specificity, and a regulatory mechanism that may be modulated by association with other proteins. Structural studies of phosphorylase kinase (Phk), the major substrate of which is glycogen phosphorylase, may be expected to shed light on its regulation. RESULTS: We report two crystal structures of the catalytic core (residues 1-298; Phk gamma trnc) of the gamma-subunit of rabbit muscle phosphorylase kinase: the binary complex with Mn2+/beta-gamma-imidoadenosine 5'-triphosphate (AMPPNP) to a resolution of 2.6 A and the binary complex with Mg2+/ADP to a resolution of 3.0 A. The structures were solved by molecular replacement using the cAMP-dependent protein kinase (cAPK) as a model. CONCLUSIONS: The overall structure of Phk gamma trnc is similar to that of the catalytic core of other protein kinases. It consists of two domians joined on one edge by a 'hinge', with the catalytic site located in the cleft between the domains. Phk gamma trnc is constitutively active, and lacks the need for an activatory phosphorylation event that is essential for many kinases. The structure exhibits an essentially 'closed' conformation of the domains which is similar to that of cAPK complexed with substrates. The phosphorylated residue that is located at the domain interface in many protein kinases and that is believed to stabilize an active conformation is substituted by a glutamate in Phk gamma trnc. The glutamate, in a similar manner to the phosphorylated residue in other protein kinases, interacts with an arginine adjacent to the catalytic aspartate but does not participate in interdomain contacts. The interactions between the enzyme and the nucleotide product of its activity, Mg2+/ADP, explain the inhibitory properties of the nucleotides that are observed in kinetic studies.
- Schulze-Gahmen U et al.
- Multiple modes of ligand recognition: crystal structures of cyclin-dependent protein kinase 2 in complex with ATP and two inhibitors, olomoucine and isopentenyladenine.
- Proteins. 1995; 22: 378-91
- Display abstract
Cyclin-dependent kinases (CDKs) are conserved regulators of the eukaryotic cell cycle with different isoforms controlling specific phases of the cell cycle. Mitogenic or growth inhibitory signals are mediated, respectively, by activation or inhibition of CDKs which phosphorylate proteins associated with the cell cycle. The central role of CDKs in cell cycle regulation makes them a potential new target for inhibitory molecules with anti-proliferative and/or anti-neoplastic effects. We describe the crystal structures of the complexes of CDK2 with a weakly specific CDK inhibitor, N6-(delta 2-isopentenyl)adenine, and a strongly specific inhibitor, olomoucine. Both inhibitors are adenine derivatives and bind in the adenine binding pocket of CDK2, but in an unexpected and different orientation from the adenine of the authentic ligand ATP. The N6-benzyl substituent in olomoucine binds outside the conserved binding pocket and is most likely responsible for its specificity. The structural information from the CDK2-olomoucine complex will be useful in directing the search for the next generation inhibitors with improved properties.
- Zhang J, Zhang F, Ebert D, Cobb MH, Goldsmith EJ
- Activity of the MAP kinase ERK2 is controlled by a flexible surface loop.
- Structure. 1995; 3: 299-307
- Display abstract
BACKGROUND: The mitogen-activated protein (MAP) kinase, ERK2, is a tightly regulated enzyme in the ubiquitous Ras-activated protein kinase cascade. ERK2 is activated by phosphorylation at two sites, Y185 and T183, that lie in the phosphorylation lip at the mouth of the catalytic site. To ascertain the role of these two residues in securing the low-activity conformation of the enzymes we have carried out crystallographic analyses and assays of phosphorylation-site mutants of ERK2. RESULTS: The crystal structures of four mutants, T183E (threonine at residue 183 is replaced by glutamate), Y185E, Y185F and the double mutant T183E/Y185E, were determined. When T183 is replaced by glutamate, few conformational changes are observed. By contrast, when Y185 is replaced by glutamate, 19 residues become disordered, including the entire phosphorylation lip and an adjacent loop. The conservative substitution of phenylalanine for Y185 also induces relatively large conformational changes. A binding site for phosphotyrosine in the active enzyme is putatively identified on the basis of the high-resolution refinement of the structure of wild-type ERK2. CONCLUSIONS: The remarkable disorder observed throughout the phosphorylation lip when Y185 is mutated shows that the stability of the phosphorylation lip is rather low. Therefore, only modest amounts of binding energy will be required to dislodge the lip for phosphorylation, and it is likely that these residues will be involved in conformational changes associated with both with binding to kinases and phosphatases and with activation. Furthermore, the low-activity structure is specifically dependent on Y185, whereas there is no such dependency on T183. Both residues, however, participate in forming the active enzyme, contributing to its tight control.
- Hubbard SR, Wei L, Ellis L, Hendrickson WA
- Crystal structure of the tyrosine kinase domain of the human insulin receptor.
- Nature. 1994; 372: 746-54
- Display abstract
The X-ray crystal structure of the tyrosine kinase domain of the human insulin receptor has been determined by multiwavelength anomalous diffraction phasing and refined to 2.1 A resolution. The structure reveals the determinants of substrate preference for tyrosine rather than serine or threonine and a novel autoinhibition mechanism whereby one of the tyrosines that is autophosphorylated in response to insulin, Tyr 1,162, is bound in the active site.
- Taylor SS, Radzio-Andzelm E
- Three protein kinase structures define a common motif.
- Structure. 1994; 2: 345-55
- Display abstract
Structural comparisons between cAMP-dependent protein kinase, cyclin-dependent kinase 2 and mitogen-activated protein kinase reveal which features are common to the protein kinase family and which are enzyme-specific.
- Zhang F, Strand A, Robbins D, Cobb MH, Goldsmith EJ
- Atomic structure of the MAP kinase ERK2 at 2.3 A resolution.
- Nature. 1994; 367: 704-11
- Display abstract
The structure of the MAP kinase ERK2, a ubiquitous protein kinase target for regulation by Ras and Raf, has been solved in its unphosphorylated low-activity conformation to a resolution of 2.3 A. The two domains of unphosphorylated ERK2 are farther apart than in the active conformation of cAMP-dependent protein kinase and the peptide-binding site is blocked by tyrosine 185, one of the two residues that are phosphorylated in the active enzyme. Activation of ERK2 is thus likely to involve both global and local conformational changes.
- Bossemeyer D, Engh RA, Kinzel V, Ponstingl H, Huber R
- Phosphotransferase and substrate binding mechanism of the cAMP-dependent protein kinase catalytic subunit from porcine heart as deduced from the 2.0 A structure of the complex with Mn2+ adenylyl imidodiphosphate and inhibitor peptide PKI(5-24).
- EMBO J. 1993; 12: 849-59
- Display abstract
The crystal structure of the porcine heart catalytic subunit of cAMP-dependent protein kinase in a ternary complex with the MgATP analogue MnAMP-PNP and a pseudosubstrate inhibitor peptide, PKI(5-24), has been solved at 2.0 A resolution from monoclinic crystals of the catalytic subunit isoform CA. The refinement is presently at an R factor of 0.194 and the active site of the molecule is well defined. The glycine-rich phosphate anchor of the nucleotide binding fold motif of the protein kinase is a beta ribbon acting as a flap with conformational flexibility over the triphosphate group. The glycines seem to be conserved to avoid steric clash with ATP. The known synergistic effects of substrate binding can be explained by hydrogen bonds present only in the ternary complex. Implications for the kinetic scheme of binding order are discussed. The structure is assumed to represent a phosphotransfer competent conformation. The invariant conserved residue Asp166 is proposed to be the catalytic base and Lys168 to stabilize the transition state. In some tyrosine kinases Lys168 is functionally replaced by an Arg displaced by two residues in the primary sequence, suggesting invariance in three-dimensional space. The structure supports an in-line transfer with a pentacoordinate transition state at the phosphorus with very few nuclear movements.
- DeBondt HL, Rosenblatt J, Jancarik J, Jones HD, Morgan DO, Kim SH
- Crystal structure of cyclin-dependent kinase 2.
- Nature. 1993; 363: 595-602
- Display abstract
Cyclin-dependent kinase 2 (CDK2) is a member of a highly conserved family of protein kinases that regulate the eukaryotic cell cycle. The crystal structures of the human CDK2 apoenzyme and its Mg2+ ATP complex have been determined to 2.4 A resolution. The structure is bi-lobate, like that of the cyclic AMP-dependent protein kinase, but contains a unique helix-loop segment that interferes with ATP and protein substrate binding and probably plays a key part in the regulation of all cyclin-dependent kinases.
- Zheng J et al.
- Crystal structure of the catalytic subunit of cAMP-dependent protein kinase complexed with MgATP and peptide inhibitor.
- Biochemistry. 1993; 32: 2154-61
- Display abstract
The structure of a ternary complex of the catalytic subunit of cAMP-dependent protein kinase, MgATP, and a 20-residue inhibitor peptide was determined at a resolution of 2.7 A using the difference Fourier technique starting from the model of the binary complex (Knighton et al., 1991a). The model of the ternary complex was refined using both X-PLOR and TNT to an R factor of 0.212 and 0.224, respectively. The orientation of the nucleotide and the interactions of MgATP with numerous conserved residues at the active site of the enzyme are clearly defined. The unique protein kinase nucleotide binding site consists of a five-stranded antiparallel beta-sheet with the base buried in a hydrophobic site along beta-strands 1 and 2 and fixed by hydrogen bonds to the N6 amino and N7 nitrogens. The small lobe secures the nucleotide via a glycine-rich loop and by ion pairing with Lys72 and Glu91. While the small lobe fixes the nontransferable alpha- and beta-phosphates in this inhibitor complex, the gamma-phosphate is secured by two Mg2+ ions and interacts both directly and indirectly with several residues in the large lobe--Asp184, Asn171, Lys168. Asp166 is positioned to serve as a catalytic base. The structure is correlated with previous chemical evidence, and the features that distinguish this nucleotide binding motif from other nucleotide binding proteins are delineated.
- Knighton DR, Zheng JH, TenEyck LF, Xuong NH, Taylor SS, Sowadski JM
- Structure of a peptide inhibitor bound to the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase.
- Science. 1991; 253: 414-20
- Display abstract
The structure of a 20-amino acid peptide inhibitor bound to the catalytic subunit of cyclic AMP-dependent protein kinase, and its interactions with the enzyme, are described. The x-ray crystal structure of the complex is the basis of the analysis. The peptide inhibitor, derived from a naturally occurring heat-stable protein kinase inhibitor, contains an amphipathic helix that is followed by a turn and an extended conformation. The extended region occupies the cleft between the two lobes of the enzyme and contains a five-residue consensus recognition sequence common to all substrates and peptide inhibitors of the catalytic subunit. The helical portion of the peptide binds to a hydrophobic groove and conveys high affinity binding. Loops from both domains converge at the active site and contribute to a network of conserved residues at the sites of magnesium adenosine triphosphate binding and catalysis. Amino acids associated with peptide recognition, nonconserved, extend over a large surface area.
- Knighton DR et al.
- Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase.
- Science. 1991; 253: 407-14
- Display abstract
The crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase complexed with a 20-amino acid substrate analog inhibitor has been solved and partially refined at 2.7 A resolution to an R factor of 0.212. The magnesium adenosine triphosphate (MgATP) binding site was located by difference Fourier synthesis. The enzyme structure is bilobal with a deep cleft between the lobes. The cleft is filled by MgATP and a portion of the inhibitor peptide. The smaller lobe, consisting mostly of amino-terminal sequence, is associated with nucleotide binding, and its largely antiparallel beta sheet architecture constitutes an unusual nucleotide binding motif. The larger lobe is dominated by helical structure with a single beta sheet at the domain interface. This lobe is primarily involved in peptide binding and catalysis. Residues 40 through 280 constitute a conserved catalytic core that is shared by more than 100 protein kinases. Most of the invariant amino acids in this conserved catalytic core are clustered at the sites of nucleotide binding and catalysis.
- Metabolism (metabolic pathways involving proteins which contain this domain)
-
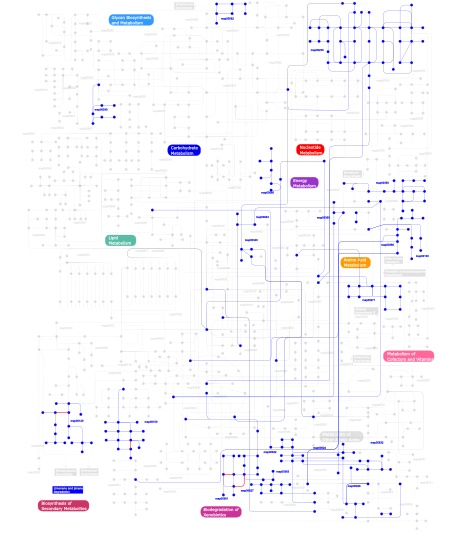
Click the image to view the interactive version of the map in iPath% proteins involved KEGG pathway ID Description 9.26  map00632
map00632Benzoate degradation via CoA ligation 9.26  map00562
map00562Inositol phosphate metabolism 4.59 map04010 MAPK signaling pathway 3.80 map04060 Cytokine-cytokine receptor interaction 3.80 map04350 TGF-beta signaling pathway 3.24 map04620 Toll-like receptor signaling pathway 3.05 map04730 Long-term depression 3.01 map04540 Gap junction 2.81 map04340 Hedgehog signaling pathway 2.65 map05212 Pancreatic cancer 2.61 map05220 Chronic myeloid leukemia 2.49 map04210 Apoptosis 2.37 map04110 Cell cycle 2.37  map00130
map00130Ubiquinone biosynthesis 2.18  map00230
map00230Purine metabolism 2.14 map04914 Progesterone-mediated oocyte maturation 2.10 map04810 Regulation of actin cytoskeleton 2.10 map05210 Colorectal cancer 1.94 map04520 Adherens junction 1.94 map04111 Cell cycle - yeast 1.82 map04920 Adipocytokine signaling pathway 1.42 map04310 Wnt signaling pathway 1.42 map04910 Insulin signaling pathway 1.42 map05215 Prostate cancer 1.38 map05221 Acute myeloid leukemia 1.27 map04012 ErbB signaling pathway 1.23 map04660 T cell receptor signaling pathway 1.19 map05213 Endometrial cancer 1.19 map04510 Focal adhesion 1.15 map04630 Jak-STAT signaling pathway 1.07 map04360 Axon guidance 1.03 map04140 Regulation of autophagy 1.03 map05214 Glioma 0.99 map05120 Epithelial cell signaling in Helicobacter pylori infection 0.95 map04720 Long-term potentiation 0.95 map04530 Tight junction 0.95 map05219 Bladder cancer 0.95 map05223 Non-small cell lung cancer 0.91 map05211 Renal cell carcinoma 0.91 map05218 Melanoma 0.87 map04150 mTOR signaling pathway 0.87 map04650 Natural killer cell mediated cytotoxicity 0.71 map04912 GnRH signaling pathway 0.67 map04664 Fc epsilon RI signaling pathway 0.59 map05222 Small cell lung cancer 0.55 map04662 B cell receptor signaling pathway 0.47 map04370 VEGF signaling pathway 0.47 map04710 Circadian rhythm 0.44 map04930 Type II diabetes mellitus 0.44 map04916 Melanogenesis 0.32 map04020 Calcium signaling pathway 0.32 map03320 PPAR signaling pathway 0.28 map05216 Thyroid cancer 0.20 map05020 Parkinson's disease 0.12  map00540
map00540Lipopolysaccharide biosynthesis 0.12 map04670 Leukocyte transendothelial migration 0.08  map00350
map00350Tyrosine metabolism 0.08 map04740 Olfactory transduction 0.08 map02020 Two-component system - General 0.08  map00150
map00150Androgen and estrogen metabolism 0.08  map00626
map00626Naphthalene and anthracene degradation 0.08 map04115 p53 signaling pathway 0.08  map00271
map00271Methionine metabolism 0.08  map00361
map00361gamma-Hexachlorocyclohexane degradation 0.08  map00360
map00360Phenylalanine metabolism 0.08  map00380
map00380Tryptophan metabolism 0.08  map00643
map00643Styrene degradation 0.08  map00622
map00622Toluene and xylene degradation 0.08  map00627
map006271,4-Dichlorobenzene degradation 0.08  map00340
map00340Histidine metabolism 0.08  map00120
map00120Bile acid biosynthesis 0.08  map00363
map00363Bisphenol A degradation 0.08  map00680
map00680Methane metabolism 0.08 map00903 Limonene and pinene degradation 0.08  map00624
map006241- and 2-Methylnaphthalene degradation 0.04 map05217 Basal cell carcinoma 0.04 map04070 Phosphatidylinositol signaling system 0.04 map04320 Dorso-ventral axis formation 0.04 map05050 Dentatorubropallidoluysian atrophy (DRPLA) This information is based on mapping of SMART genomic protein database to KEGG orthologous groups. Percentage points are related to the number of proteins with STYKc domain which could be assigned to a KEGG orthologous group, and not all proteins containing STYKc domain. Please note that proteins can be included in multiple pathways, ie. the numbers above will not always add up to 100%.
- Structure (3D structures containing this domain)
3D Structures of STYKc domains in PDB
PDB code Main view Title 1b6c 
CRYSTAL STRUCTURE OF THE CYTOPLASMIC DOMAIN OF THE TYPE I TGF-BETA RECEPTOR IN COMPLEX WITH FKBP12 1cki 
RECOMBINANT CASEIN KINASE I DELTA TRUNCATION MUTANT CONTAINING RESIDUES 1-317 1ckj 
CASEIN KINASE I DELTA TRUNCATION MUTANT CONTAINING RESIDUES 1-317 COMPLEX WITH BOUND TUNGSTATE 1csn 
BINARY COMPLEX OF CASEIN KINASE-1 WITH MGATP 1eh4 
BINARY COMPLEX OF CASEIN KINASE-1 FROM S. POMBE WITH AN ATP COMPETITIVE INHIBITOR, IC261 1ias 
CYTOPLASMIC DOMAIN OF UNPHOSPHORYLATED TYPE I TGF-BETA RECEPTOR CRYSTALLIZED WITHOUT FKBP12 1py5 
Crystal Structure of TGF-beta receptor I kinase with inhibitor 1rw8 
Crystal Structure of TGF-beta receptor I kinase with ATP site inhibitor 1uwh 
The complex of wild type B-RAF and BAY439006 1uwj 
The complex of mutant V599E B-RAF and BAY439006 1vjy 
Crystal Structure of a Naphthyridine Inhibitor of Human TGF-beta Type I Receptor 1wak 
X-ray structure of SRPK1 1wbp 
SRPK1 bound to 9mer docking motif peptide 1x8b 
Structure of human Wee1A kinase: kinase domain complexed with inhibitor PD0407824 2a19 
PKR kinase domain- eIF2alpha- AMP-PNP complex. 2a1a 
PKR kinase domain-eIF2alpha Complex 2buj 
Crystal structure of the human Serine-threonine Kinase 16 in complex with staurosporine 2c47 
Structure of casein kinase 1 gamma 2 2chl 
Structure of casein kinase 1 gamma 3 2cmw 
Structure of Human Casein kinase 1 gamma-1 in complex with 2-(2- Hydroxyethylamino)-6-(3-chloroanilino)-9-isopropylpurine (CASP TARGET) 2csn 
BINARY COMPLEX OF CASEIN KINASE-1 WITH CKI7 2eva 
Structural Basis for the Interaction of TAK1 Kinase with its Activating Protein TAB1 2fb8 
Structure of the B-Raf kinase domain bound to SB-590885 2h34 
Apoenzyme crystal structure of the tuberculosis serine/threonine kinase, PknE 2in6 
Wee1 kinase complex with inhibitor PD311839 2io6 
Wee1 kinase complexed with inhibitor PD330961 2izr 
Structure of casein kinase gamma 3 in complex with inhibitor 2izs 
Structure of casein kinase gamma 3 in complex with inhibitor 2izt 
Structure of casein kinase gamma 3 in complex with inhibitor 2izu 
Structure of casein kinase gamma 3 in complex with inhibitor 2jii 
Structure of vaccinia related kinase 3 2kty 
Solution Structure of human Vaccinia Related Kinase-1 2kul 
Solution structure of human vaccinia related kinase 1(VRK1) 2lav 
NMR solution structure of human Vaccinia-Related Kinase 1 2nru 
Crystal structure of IRAK-4 2nry 
Crystal structure of IRAK-4 2o8y 
Apo IRAK4 Kinase Domain 2oib 
Crystal structure of IRAK4 kinase domain apo form 2oic 
Crystal structure of IRAK4 kinase domain complexed with staurosporine 2oid 
Crystal structure of IRAK4 kinase domain complexed with AMPPNP 2pml 
Crystal structure of PfPK7 in complex with an ATP analogue 2pmn 
Crystal structure of PfPK7 in complex with an ATP-site inhibitor 2pmo 
Crystal structure of PfPK7 in complex with hymenialdisine 2pzi 
Crystal Structure of Protein kinase PknG from Mycobacterium tuberculosis in Complex with Tetrahydrobenzothiophene AX20017 2qkw 
Structural basis for activation of plant immunity by bacterial effector protein AvrPto 2qlu 
Crystal structure of Activin receptor type II kinase domain from human 2rio 
Structure of the dual enzyme Ire1 reveals the basis for catalysis and regulation of non-conventional splicing 2rsv 
Solution structure of human full-length vaccinia related kinase 1 (VRK1) 2v62 
Structure of vaccinia-related kinase 2 2vuw 
Structure of human haspin kinase domain 2vwb 
Structure of the archaeal Kae1-Bud32 fusion protein MJ1130: a model for the eukaryotic EKC-KEOPS subcomplex involved in transcription and telomere homeostasis. 2w1z 
ROP2 from Toxoplasma gondii: A virulence factor with a protein- kinase fold and no enzymatic activity. 2wb8 
Crystal structure of Haspin kinase 2wot 
ALK5 IN COMPLEX WITH 4-((5,6-dimethyl-2-(2-pyridyl)-3-pyridyl)oxy)-N-( 3,4,5-trimethoxyphenyl)pyridin-2-amine 2wou 
ALK5 IN COMPLEX WITH 4-((4-((2,6-dimethyl-3-pyridyl)oxy)-2-pyridyl) amino)benzenesulfonamide 2wtk 
Structure of the heterotrimeric LKB1-STRADalpha-MO25alpha complex 2x7g 
Structure of human serine-arginine-rich protein-specific kinase 2 ( SRPK2) bound to purvalanol B 2x7o 
Crystal structure of TGFbRI complexed with an indolinone inhibitor 2y4i 
KSR2-MEK1 heterodimer 2yiy 
Crystal structure of compound 8 bound to TAK1-TAB 2z2w 
Humand Wee1 kinase complexed with inhibitor PF0335770 3beg 
Crystal structure of SR protein kinase 1 complexed to its substrate ASF/SF2 3bi6 
Wee1 kinase complex with inhibitor PD352396 3biz 
Wee1 kinase complex with inhibitor PD331618 3byv 
Crystal structure of Toxoplasma gondii specific rhoptry antigen kinase domain 3c4c 
B-Raf Kinase in Complex with PLX4720 3cqe 
Wee1 kinase complex with inhibitor PD074291 3cr0 
Wee1 kinase complex with inhibitor PD259_809 3d4q 
Pyrazole-based inhibitors of B-Raf kinase 3dlz 
Crystal structure of human haspin in complex with AMP 3dzo 
Crystal structure of a rhoptry kinase from toxoplasma gondii 3e7v 
Crystal Structure of Human Haspin with a pyrazolo-pyrimidine ligand 3en9 
Structure of the Methanococcus jannaschii KAE1-BUD32 fusion protein 3enh 
Crystal structure of Cgi121/Bud32/Kae1 complex 3f2n 
Crystal Structure of Human Haspin with an Imidazo-pyridazine ligand 3faa 
Crystal structure of TGFbRI complexed with a 2-aminoimidazole inhibitor 3fbv 
Crystal structure of the oligomer formed by the kinase-ribonuclease domain of Ire1 3fmd 
Crystal Structure of Human Haspin with an Isoquinoline ligand 3fpq 
Crystal structure of the kinase domain of WNK1 3g2f 
Crystal structure of the kinase domain of bone morphogenetic protein receptor type II (BMPR2) at 2.35 A resolution 3gni 
Structure of STRAD and MO25 3gxl 
ALK-5 kinase complex with GW857175 3h9r 
Crystal structure of the kinase domain of type I activin receptor (ACVR1) in complex with FKBP12 and dorsomorphin 3hgk 
crystal structure of effect protein AvrptoB complexed with kinase Pto 3hmm 
Structure of Alk5 + GW855857 3idp 
B-Raf V600E kinase domain in complex with an aminoisoquinoline inhibitor 3ii5 
The Complex of wild-type B-RAF with Pyrazolo pyrimidine inhibitor 3iq7 
Crystal Structure of human Haspin in complex with 5-Iodotubercidin 3kcf 
Crystal structure of TGFbRI complexed with a pyrazolone inhibitor 3kmu 
Crystal structure of the ILK/alpha-parvin core complex (apo) 3kmw 
Crystal structure of the ILK/alpha-parvin core complex (MgATP) 3lj0 
IRE1 complexed with ADP and Quercetin 3lj1 
IRE1 complexed with Cdk1/2 Inhibitor III 3lj2 
IRE1 complexed with JAK Inhibitor I 3mdy 
Crystal structure of the cytoplasmic domain of the bone morphogenetic protein receptor type-1B (BMPR1B) in complex with FKBP12 and LDN-193189 3mtf 
Crystal structure of the ACVR1 kinase in complex with a 2-aminopyridine inhibitor 3my0 
Crystal structure of the ACVRL1 (ALK1) kinase domain bound to LDN-193189 3og7 
B-Raf Kinase V600E oncogenic mutant in complex with PLX4032 3omv 
Crystal structure of c-raf (raf-1) 3oom 
Crystal structure of the ACVR1 kinase domain in complex with the imidazo[1,2-b]pyridazine inhibitor K00507 3op5 
Human vaccinia-related kinase 1 3orm 
Mycobacterium tuberculosis PknB kinase domain D76A mutant 3p1a 
Structure of human Membrane-associated Tyrosine- and Threonine-specific cdc2-inhibitory kinase MYT1 (PKMYT1) 3p23 
Crystal structure of the Human kinase and RNase domains in complex with ADP 3p86 
Crystal structure of CTR1 kinase domain mutant D676N in complex with staurosporine 3ppj 
Human B-Raf Kinase in Complex with a Furopyridine Inhibitor 3ppk 
Human B-Raf Kinase in Complex with a Non-Oxime Furopyridine Inhibitor 3prf 
Crystal Structure of Human B-Raf Kinase Domain in Complex with a Non-Oxime Furopyridine Inhibitor 3pri 
Crystal Structure of Human B-Raf Kinase in Complex with a Non-Oxime Furopyridine Inhibitor 3psb 
Furo[2,3-c]pyridine-based Indanone Oximes as Potent and Selective B-Raf Inhibitors 3psd 
Non-oxime pyrazole based inhibitors of B-Raf kinase 3q4c 
Crystal Structure of Wild Type BRAF kinase domain in complex with organometallic inhibitor CNS292 3q4t 
Crystal structure of Activin receptor type-IIA (ACVR2A) kinase domain in complex with dorsomorphin 3q4u 
Crystal structure of the ACVR1 kinase domain in complex with LDN-193189 3q5z 
Crystal structure of virulent allele ROP5B pseudokinase domain 3q60 
Crystal structure of virulent allele ROP5B pseudokinase domain bound to ATP 3q96 
B-Raf kinase domain in complex with a tetrahydronaphthalene inhibitor 3qa8 
Crystal Structure of inhibitor of kappa B kinase beta 3qd2 
Crsytal structure of mouse PERK kinase domain 3rep 
Crystal structure of the ILK/alpha-parvin core complex (MnATP) 3rzf 
Crystal Structure of Inhibitor of kappaB kinase beta (I4122) 3s95 
Crystal structure of the human LIMK1 kinase domain in complex with staurosporine 3sdj 
Structure of RNase-inactive point mutant of oligomeric kinase/RNase Ire1 3sdm 
Structure of oligomeric kinase/RNase Ire1 in complex with an oligonucleotide 3skc 
Human B-Raf Kinase in Complex with an Amide Linked Pyrazolopyridine Inhibitor 3soc 
Crystal structure of Activin receptor type-IIA (ACVR2A) kinase domain in complex with a quinazolin 3sv0 
Crystal structure of casein kinase-1 like protein in plant 3tl8 
The AvrPtoB-BAK1 complex reveals two structurally similar kinaseinteracting domains in a single type III effector 3tv4 
Human B-Raf Kinase Domain in Complex with an Bromopyridine Benzamide Inhibitor 3tv6 
Human B-Raf Kinase Domain in Complex with a Methoxypyrazolopyridinyl Benzamide Inhibitor 3tzm 
TGF-beta Receptor type 1 in complex with SB431542 3uim 
Structural basis for the impact of phosphorylation on plant receptor-like kinase BAK1 activation 3uiu 
Crystal structure of Apo-PKR kinase domain 3ulz 
Crystal structure of apo BAK1 3uys 
Crystal structure of apo human ck1d 3uyt 
crystal structure of ck1d with PF670462 from P1 crystal form 3uzp 
crystal structure of ck1d with PF670462 from P21 crystal form 3zon 
Human TYK2 pseudokinase domain bound to a kinase inhibitor 4asx 
Crystal structure of Activin receptor type-IIA (ACVR2A) kinase domain in complex with a beta-carboline inhibitor 4ax8 
Medium resolution structure of the bifunctional kinase- methyltransferase WbdD 4azs 
High resolution (2.2 A) crystal structure of WbdD. 4azt 
Co-crystal structure of WbdD and kinase inhibitor LY294002. 4azv 
Co-crystal structure of WbdD and kinase inhibitor GW435821x. 4azw 
Crystal structure of monomeric WbdD. 4bgg 
Crystal structure of the ACVR1 kinase in complex with LDN-213844 4btf 
Structure of MLKL 4btj 
TTBK1 in complex with ATP 4btk 
TTBK1 in complex with inhibitor 4btm 
TTBK1 in complex with inhibitor 4bwk 
Structure of Neurospora crassa PAN3 pseudokinase 4bwp 
Structure of Drosophila Melanogaster PAN3 pseudokinase 4bwx 
Structure of Neurospora crassa PAN3 pseudokinase mutant 4c02 
Crystal structure of human ACVR1 (ALK2) in complex with FKBP12.6 and dorsomorphin 4c57 
Structure of GAK kinase in complex with a nanobody 4c58 
Structure of GAK kinase in complex with nanobody (NbGAK_4) 4c59 
Structure of GAK kinase in complex with nanobody (NbGAK_4) 4c8b 
Structure of the kinase domain of human RIPK2 in complex with ponatinib 4ci6 
4CI6 4cqe 
4CQE 4cyi 
4CYI 4cyj 
4CYJ 4czy 
4CZY 4dbn 
Crystal Structure of the Kinase domain of Human B-raf with a [1,3]thiazolo[5,4-b]pyridine derivative 4dn5 
Crystal Structure of NF-kB-inducing Kinase (NIK) 4dym 
Crystal structure of the ACVR1 kinase domain in complex with the imidazo[1,2-b]pyridazine inhibitor K00135 4e26 
BRAF in complex with an organic inhibitor 7898734 4e3c 
X-ray crystal structure of human IKK2 in an active conformation 4e4x 
Crystal Structure of B-Raf Kinase Domain in Complex with a Dihydropyrido[2,3-d]pyrimidinone-based Inhibitor 4ehe 
B-Raf Kinase Domain in Complex with an Aminothienopyrimidine-based Inhibitor 4ehg 
B-Raf Kinase Domain in Complex with an Aminopyridimine-based Inhibitor 4eut 
Structure of BX-795 Complexed with Unphosphorylated Human TBK1 Kinase-ULD Domain 4euu 
Structure of BX-795 Complexed with Human TBK1 Kinase Domain Phosphorylated on Ser172 4f0f 
Crystal Structure of the Roco4 Kinase Domain bound to AppCp from D. discoideum 4f0g 
Crystal Structure of the Roco4 Kinase Domain from D. discoideum 4f1m 
Crystal Structure of the G1179S Roco4 Kinase Domain bound to AppCp from D. discoideum. 4f1o 
Crystal Structure of the L1180T mutant Roco4 Kinase Domain from D. discoideum bound to AppCp 4f1t 
Crystal Structure of the Roco4 Kinase Domain from D. discoideum bound to the ROCK Inhibitor H1152 4f99 
Human CDC7 kinase in complex with DBF4 and nucleotide 4f9a 
Human CDC7 kinase in complex with DBF4 and nucleotide 4f9b 
Human CDC7 kinase in complex with DBF4 and PHA767491 4f9c 
Human CDC7 kinase in complex with DBF4 and XL413 4fc0 
Crystal Structure of Human Kinase Domain of B-raf with a DFG-out Inhibitor 4fi1 
Crystal structure of scCK2 alpha in complex with ATP 4fk3 
B-Raf Kinase V600E Oncogenic Mutant in Complex with PLX3203 4fvp 
Crystal structure of the Jak2 pseudokinase domain (apo form) 4fvq 
Crystal structure of the Jak2 pseudokinase domain (Mg-ATP-bound form) 4fvr 
Crystal structure of the Jak2 pseudokinase domain mutant V617F (Mg-ATP-bound form) 4g16 
Crystal structure of ck1g3 with 2-[(4-{[3-(TRIFLUOROMETHYL)PYRIDIN2-YL]OXY}PHENYL)AMINO]-1H-BENZIMIDAZOLE-6-CARBONITRILE 4g17 
Crystal structure of ck1g3 with 2-[(4-TERT-BUTYLPHENYL)AMINO]-1H-BENZIMIDAZOLE-6-CARBONITRILE 4g31 
Crystal Structure of GSK6414 Bound to PERK (R587-R1092, delete A660-T867) at 2.28 A Resolution 4g34 
Crystal Structure of GSK6924 Bound to PERK (R587-R1092, delete A660-T867) at 2.70 A Resolution 4g3c 
Crystal structure of apo murine Nf-kappaB inducing kinase (NIK) 4g3d 
Crystal structure of human NF-kappaB inducing kinase (NIK) 4g3e 
Crystal structure of murine NF-kappaB inducing kinase (NIK) bound to a 6-alkynylindoline (cmp1) 4g3f 
Crystal structure of murine NF-kappaB inducing kinase (NIK) bound to a 2-(aminothiazoly)phenol (cmp2) 4g3g 
Crystal structure of murine NF-kappaB inducing kinase (NIK) V408L bound to a 2-(aminothiazolyl)phenol (cmp3) 4g9c 
Human B-Raf Kinase Domain bound to a Type II Pyrazolopyridine Inhibitor 4g9r 
B-Raf V600E Kinase Domain Bound to a Type II Dihydroquinazoline Inhibitor 4gs6 
Irreversible Inhibition of TAK1 Kinase by 5Z-7-Oxozeaenol 4h58 
BRAF in complex with compound 3 4hgl 
Crystal structure of ck1g3 with compound 1 4hgs 
Crystal structure of ck1gs with compound 13 4hgt 
Crystal structure of ck1d with compound 13 4hnf 
Crystal structure of ck1d in complex with pf4800567 4hni 
crystal structure of ck1e in complex with PF4800567 4hok 
crystal structure of apo ck1e 4i92 
Structure of the BSK8 kinase domain 4i93 
Structure of the BSK8 kinase domain (SeMet labeled) 4i94 
Structure of BSK8 in complex with AMP-PNP 4idt 
Crystal Structure of NIK with 11-bromo-5,6,7,8-tetrahydropyrimido[4',5':3,4]cyclohepta[1,2-b]indol-2-amine (T28) 4idv 
Crystal Structure of NIK with compound 4-{3-[2-amino-5-(2-methoxyethoxy)pyrimidin-4-yl]-1H-indol-5-yl}-2-methylbut-3-yn-2-ol (13V) 4im0 
Structure of Tank-Binding Kinase 1 4im2 
Structure of Tank-Binding Kinase 1 4im3 
Structure of Tank-Binding Kinase 1 4ith 
Crystal structure of RIP1 kinase in complex with necrostatin-1 analog 4iti 
Crystal structure of RIP1 kinase in complex with necrostatin-3 analog 4itj 
Crystal structure of RIP1 kinase in complex with necrostatin-4 4iw0 
Crystal structure and mechanism of activation of TBK1 4iwo 
Crystal structure and mechanism of activation of TBK1 4iwp 
Crystal structure and mechanism of activation of TBK1 4iwq 
Crystal structure and mechanism of activation of TBK1 4ix3 
Crystal structure of a Stt7 homolog from Micromonas algae 4ix4 
Crystal structure of a Stt7 homolog from Micromonas algae in complex with ADP 4ix5 
Crystal structure of a Stt7 homolog from Micromonas algae in complex with AMP-PNP 4ix6 
Crystal structure of a Stt7 homolog from Micromonas algae soaked with ATP 4jjr 
A P21 crystal form of mammalian casein kinase 1 delta 4jl9 
Crystal structure of mouse TBK1 bound to BX795 4jlc 
Crystal structure of mouse TBK1 bound to SU6668 4jqe 
Crystal structure of scCK2 alpha in complex with AMPPN 4jr7 
Crystal structure of scCK2 alpha in complex with GMPPNP 4jrn 
ROP18 kinase domain in complex with AMP-PNP and sucrose 4jvg 
B-Raf Kinase in Complex with Birb796 4kb8 
CK1d in complex with 1-{4-[3-(4-FLUOROPHENYL)-1-METHYL-1H-PYRAZOL-4-YL]PYRIDIN-2-YL}-N-METHYLMETHANAMINE ligand 4kba 
CK1d in complex with 9-[3-(4-fluorophenyl)-1-methyl-1H-pyrazol-4-yl]-2,3,4,5-tetrahydropyrido[2,3-f][1,4]oxazepine inhibitor 4kbc 
CK1d in complex with {4-[3-(4-FLUOROPHENYL)-1H-PYRAZOL-4-YL]PYRIDIN-2-YL}METHANOL inhibitor 4kbk 
CK1d in complex with (3S)-3-{4-[3-(4-fluorophenyl)-1-methyl-1H-pyrazol-4-yl]pyridin-2-yl}morpholine inhibitor 4kik 
Human IkB kinase beta 4ksp 
Crystal Structure of Human B-raf bound to a DFG-out Inhibitor TAK-632 4ksq 
Crystal Structure of Human B-raf bound to a DFG-out Inhibitor 5B 4l00 
Crystal structure of the apo Jak1 pseudokinase domain 4l01 
Crystal structure of the V658F apo Jak1 pseudokinase domain 4l3p 
Crystal Structure of 2-(1-benzothiophen-7-yl)-4-[1-(piperidin-4-yl)-1H-pyrazol-4-yl]furo[2,3-c]pyridin-7-amine bound to TAK1-TAB1 4l52 
Crystal Structure of 1-(4-{4-[7-amino-2-(1,2,3-benzothiadiazol-7-yl)furo[2,3-c]pyridin-4-yl]-1H-pyrazol-1-yl}piperidin-1-yl)ethan-1-one bound to TAK1-TAB1 4l53 
Crystal Structure of (1R,4R)-4-{4-[7-amino-2-(1,2,3-benzothiadiazol-7-yl)-3-chlorofuro[2,3-c]pyridin-4-yl]-1H-pyrazol-1-yl}cyclohexan-1-ol bound to TAK1-TAB1 4l68 
Structure of the psedudokinase domain of BIR2, an immune regulator of the RLK/Pelle family 4lfi 
Crystal structure of scCK2 alpha in complex with GMPPNP 4lv5 
Murine IRGa6 bound to Toxoplasma ROP5B, a pseudokinase GDI 4lv8 
Murine IRGa6 bound to Toxoplasma ROP5C, a pseudokinase GDI 4m66 
Crystal structure of the mouse RIP3 kinase domain 4m67 
Crystal structure of the human MLKL kinase-like domain 4m68 
Crystal structure of the mouse MLKL kinase-like domain 4m69 
Crystal structure of the mouse RIP3-MLKL complex 4m7i 
4M7I 4mbj 
Human B-Raf Kinase Domain in Complex with an Imidazopyridine-based Inhibitor 4mne 
4MNE 4mnf 
4MNF 4mwh 
Crystal structure of scCK2 alpha in complex with ATP 4mwi 
Crystal structure of the human MLKL pseudokinase domain 4neu 
X-ray structure of Receptor Interacting Protein 1 (RIP1)kinase domain with a 1-aminoisoquinoline inhibitor 4nfm 
Human tau tubulin kinase 1 (TTBK1) 4nfn 
Human tau tubulin kinase 1 (TTBK1) complexed with 3-({5-[(4-amino-4-methylpiperidin-1-yl)methyl]pyrrolo[2,1-f][1,2,4]triazin-4-yl}amino)-5-bromophenol 4nt4 
Crystal structure of the kinase domain of Gilgamesh isoform I from Drosophila melanogaster 4o1o 
Crystal Structure of RNase L in complex with 2-5A 4o1p 
Crystal Structure of RNase L in complex with 2-5A and AMP-PNP 4o38 
Crystal structure of the human cyclin G associated kinase (GAK) 4o91 
4O91 4oau 
Complete human RNase L in complex with biological activators. 4oav 
Complete human RNase L in complex with 2-5A (5'-ppp heptamer), AMPPCP and RNA substrate. 4oh4 
4OH4 4oli 
4OLI 4ouc 
Structure of human haspin in complex with histone H3 substrate 4ow8 
4OW8 4pl3 
4PL3 4pl4 
4PL4 4pl5 
4PL5 4pp7 
Highly Potent and Selective 3-N-methylquinazoline-4(3H)-one Based Inhibitors of B-RafV600E Kinase 4pwn 
4PWN 4q2a 
4Q2A 4q5j 
4Q5J 4qpm 
4QPM 4qtc 
4QTC 4r5y 
4R5Y 4r8q 
4R8Q 4rmz 
4RMZ 4rzv 
4RZV 4rzw 
4RZW 4tn6 
4TN6 4tpt 
4TPT 4tw9 
4TW9 4twc 
4TWC 4u6r 
4U6R 4u97 
4U97 4u9a 
4U9A 4uw0 
4UW0 4w9w 
4W9W 4w9x 
4W9X 4wo5 
4WO5 4wov 
4WOV 4wsq 
4WSQ 4wua 
4WUA 4ww5 
4WW5 4ww7 
4WW7 4ww9 
4WW9 4wwa 
4WWA 4x0m 
4X0M 4x2f 
4X2F 4x2g 
4X2G 4x2j 
4X2J 4x2k 
4X2K 4x2n 
4X2N 4x3f 
4X3F 4x7h 
4X7H 4x7j 
4X7J 4x7k 
4X7K 4x7l 
4X7L 4x7n 
4X7N 4x7o 
4X7O 4xh0 
4XH0 4xhg 
4XHG 4xhh 
4XHH 4xhl 
4XHL 4xr7 
4XR7 4xs2 
4XS2 4xv1 
4XV1 4xv2 
4XV2 4xv3 
4XV3 4xv9 
4XV9 4y0x 
4Y0X 4y12 
4Y12 4y73 
4Y73 4y83 
4Y83 4y85 
4Y85 4y8d 
4Y8D 4yff 
4YFF 4yfi 
4YFI 4yht 
4YHT 4yo6 
4YO6 4yp8 
4YP8 4yz9 
4YZ9 4yzc 
4YZC 4yzd 
4YZD 4yzm 
4YZM 4yzn 
4YZN 4z7g 
4Z7G 4z7h 
4Z7H 4ztl 
4ZTL 4ztm 
4ZTM 4ztn 
4ZTN 5ar2 
5AR2 5ar3 
5AR3 5ar4 
5AR4 5ar5 
5AR5 5ar7 
5AR7 5ar8 
5AR8 5c01 
5C01 5c03 
5C03 5c9c 
5C9C 5ce3 
5CE3 5cek 
5CEK 5cem 
5CEM 5cen 
5CEN 5ceo 
5CEO 5cep 
5CEP 5ceq 
5CEQ 5ckw 
5CKW 5clr 
5CLR 5csw 
5CSW 5csx 
5CSX 5ct7 
5CT7 5cyz 
5CYZ 5czo 
5CZO 5dfz 
5DFZ 5dmz 
5DMZ 5drb 
5DRB 5e7r 
5E7R 5e8s 
5E8S 5e8t 
5E8T 5e8u 
5E8U 5e8v 
5E8V 5e8w 
5E8W 5e8x 
5E8X 5e8y 
5E8Y 5e8z 
5E8Z 5e90 
5E90 5e91 
5E91 5e92 
5E92 5ebz 
5EBZ 5fd2 
5FD2 5fqd 
5FQD 5fri 
5FRI 5gjd 
5GJD 5gjf 
5GJF 5gjg 
5GJG 5hes 
5HES 5hgi 
5HGI 5hi2 
5HI2 5hid 
5HID 5hie 
5HIE 5htb 
5HTB 5htc 
5HTC 5hvj 
5HVJ 5hvk 
5HVK 5hx6 
5HX6 5i3o 
5I3O 5i3r 
5I3R 5i4n 
5I4N 5ih4 
5IH4 5ih5 
5IH5 5ih6 
5IH6 5ikw 
5IKW 5ita 
5ITA 5iu2 
5IU2 5j0a 
5J0A 5j79 
5J79 5j7b 
5J7B 5jga 
5JGA 5jgb 
5JGB 5jgd 
5JGD 5jrq 
5JRQ 5jsm 
5JSM 5jt2 
5JT2 5kc2 
5KC2 5khu 
5KHU 5kkr 
5KKR 5knj 
5KNJ 5ko1 
5KO1 5kx7 
5KX7 5kx8 
5KX8 5l4q 
5L4Q 5l6w 
5L6W 5lpb 
5LPB 5lpv 
5LPV 5lpw 
5LPW 5lpy 
5LPY 5lpz 
5LPZ 5te0 
5TE0 5tf9 
5TF9 5tkx 
5TKX 5tqw 
5TQW 5tqx 
5TQX 5tqy 
5TQY

