The domain within your query sequence starts at position 709 and ends at position 755; the E-value for the C1 domain shown below is 3.15e-8.
RTHRLRKLRTPAKCRECNSYVYFQGAECEECCLACHKKCLETLAIQC
C1Protein kinase C conserved region 1 (C1) domains (Cysteine-rich domains) |
|---|
| SMART accession number: | SM00109 |
|---|---|
| Description: | Some bind phorbol esters and diacylglycerol. Some bind RasGTP. Zinc-binding domains. |
| Interpro abstract (IPR002219): | Diacylglycerol (DAG) is an important second messenger. Phorbol esters (PE) are analogues of DAG and potent tumour promoters that cause a variety of physiological changes when administered to both cells and tissues. DAG activates a family of serine/threonine protein kinases, collectively known as protein kinase C (PKC) [ (PUBMED:1396661) ]. Phorbol esters can directly stimulate PKC. The N-terminal region of PKC, known as C1, has been shown [ (PUBMED:2500657) ] to bind PE and DAG in a phospholipid and zinc-dependent fashion. The C1 region contains one or two copies (depending on the isozyme of PKC) of a cysteine-rich domain, which is about 50 amino-acid residues long, and which is essential for DAG/PE-binding. The DAG/PE-binding domain binds two zinc ions; the ligands of these metal ions are probably the six cysteines and two histidines that are conserved in this domain. |
| GO process: | intracellular signal transduction (GO:0035556) |
| Family alignment: |
There are 58922 C1 domains in 42393 proteins in SMART's nrdb database.
Click on the following links for more information.
- Evolution (species in which this domain is found)
-
Taxonomic distribution of proteins containing C1 domain.
This tree includes only several representative species. The complete taxonomic breakdown of all proteins with C1 domain is also avaliable.
Click on the protein counts, or double click on taxonomic names to display all proteins containing C1 domain in the selected taxonomic class.
- Cellular role (predicted cellular role)
-
Binding / catalysis: diacylglycerol-binding, phorbol-ester binding, zinc-binding, RasGTP-binding
- Literature (relevant references for this domain)
-
Primary literature is listed below; Automatically-derived, secondary literature is also avaliable.
- Hurley JH, Newton AC, Parker PJ, Blumberg PM, Nishizuka Y
- Taxonomy and function of C1 protein kinase C homology domains.
- Protein Sci. 1997; 6: 477-80
- Display abstract
C1 domains are compact alpha/beta structural units of about 50 amino acids which tightly bind two zinc ions. These domains were first discovered as the loci of phorbol ester and diacylglycerol binding to conventional protein kinase C isozymes, which contain 2 C1 domains (C1A and C1B) in their N-terminal regulatory regions. We present a comprehensive list of 54 C1 domains occurring singly or doubly in 34 different proteins. Many C1 domains and C1 domain-containing proteins bind phorbol esters, but many others do not. By combining analysis of 54 C1 domain sequences with information from previously reported solution and crystal structure determinations and site-directed mutagenesis, profiles are derived and used to classify C1 domains. Twenty-six C1 domains fit the profile for phorbol-ester binding and are termed "typical." Twenty-eight other domains fit the profile for the overall C1 domain fold but do not fit the profile for phorbol ester binding, and are termed "atypical." Proteins containing typical C1 domains are predicted to be regulated by diacylglycerol, whereas those containing only atypical domains are not.
- Xu RX, Pawelczyk T, Xia TH, Brown SC
- NMR structure of a protein kinase C-gamma phorbol-binding domain and study of protein-lipid micelle interactions.
- Biochemistry. 1997; 36: 10709-17
- Display abstract
Classical protein kinase C (PKC) family members are activated by the binding of various ligands to one of several cysteine-rich domains of the enzyme. The natural agonist, diacylglycerol (DAG), and the natural product superagonist, phorbol dibutyrate (PDB), activate the enzyme to produce wide-ranging physiological effects. The second cysteine-rich (Cys2) domain of rat brain PKC-gamma was expressed and labeled with 15N and 13C, and the solution structure was determined to high resolution using multidimensional heteronuclear NMR methods. The phorbol binding site was identified by titrating this domain with phorbol-12,13-dibutyrate (PDB) in the presence of organic cosolvents. Titrations of this domain with lipid micelles, in the absence and presence of phorbols, indicate selective broadening of some resonances. The observed behavior indicates conformational exchange between bound and free states upon protein-micelle interaction. The data also suggest that half of the domain, including the phorbol site and one of the zinc sites, is capable of inserting into membranes.
- Mott HR, Carpenter JW, Zhong S, Ghosh S, Bell RM, Campbell SL
- The solution structure of the Raf-1 cysteine-rich domain: a novel ras and phospholipid binding site.
- Proc Natl Acad Sci U S A. 1996; 93: 8312-7
- Display abstract
The Raf-1 protein kinase is the best-characterized downstream effector of activated Ras. Interaction with Ras leads to Raf-1 activation and results in transduction of cell growth and differentiation signals. The details of Raf-1 activation are unclear, but our characterization of a second Ras-binding site in the cysteine-rich domain (CRD) and the involvement of both Ras-binding sites in effective Raf-1-mediated transformation provides insight into the molecular aspects and consequences of Ras-Raf interactions. The Raf-1 CRD is a member of an emerging family of domains, many of which are found within signal transducing proteins. Several contain binding sites for diacylglycerol (or phorbol esters) and phosphatidylserine and are believed to play a role in membrane translocation and enzyme activation. The CRD from Raf-1 does not bind diacylglycerol but interacts with Ras and phosphatidylserine. To investigate the ligand-binding specificities associated with CRDs, we have determined the solution structure of the Raf-1 CRD using heteronuclear multidimensional NMR. We show that there are differences between this structure and the structures of two related domains from protein kinase C (PKC). The differences are confined to regions of the CRDs involved in binding phorbol ester in the PKC domains. Since phosphatidylserine is a common ligand, we expect its binding site to be located in regions where the structures of the Raf-1 and PKC domains are similar. The structure of the Raf-1 CRD represents an example of this family of domains that does not bind diacylglycerol and provides a framework for investigating its interactions with other molecules.
- Palmer RH, Ridden J, Parker PJ
- Cloning and expression patterns of two members of a novel protein-kinase-C-related kinase family.
- Eur J Biochem. 1995; 227: 344-51
- Display abstract
The cDNA clones for two members of a novel protein kinase family were isolated and sequenced. These protein-kinase-C-related kinases, PRK1 and PRK2, display extensive identity to each other, revealing non-kinase domain similar regions. HR1 and HR2. HR1 contains a motif repeated three times (HR1a-c), while HR2 shows similarity to the amino-terminal sequence of protein-kinase-C epsilon and eta isotypes. Both PRK1 and PRK2, expressed in COS 1 cells, are autophosphorylated in immunoprecipitates, indicating intrinsic kinase activity. PRK1 and PRK2, as well as a third member of this family, PRK3, show distinct patterns of expression in adult tissues.
- Hommel U, Zurini M, Luyten M
- Solution structure of a cysteine rich domain of rat protein kinase C.
- Nat Struct Biol. 1994; 1: 383-7
- Display abstract
Intracellular protein phosphorylation by protein kinase C (PKC) plays a major role in the translation of extracellular signals into cellular events. Speculations on the structural basis for PKC activation are based on sequence homology between their cysteine-rich domains (CRD) and the DNA-binding 'zinc-fingers'. We produced a fragment comprising the second CRD (CRD2) of rat PKC-alpha and determined its three-dimensional structure in solution by NMR spectroscopy. This revealed that CRD2 adopts a globular fold allowing two non-consecutive sets of zinc-binding residues to form two separate metal-binding sites. The fold is different to those previously proposed and allows insight into the molecular topology of a family of homologous proteins.
- Hall C et al.
- Novel human brain cDNA encoding a 34,000 Mr protein n-chimaerin, related to both the regulatory domain of protein kinase C and BCR, the product of the breakpoint cluster region gene.
- J Mol Biol. 1990; 211: 11-6
- Display abstract
A novel human brain complementary DNA sequence encodes n-chimaerin, a 34,000 Mr protein. A single cysteine-rich sequence CX2CX13CX2CX7CX7C in the N-terminal half of n-chimaerin shares almost 50% identity with corresponding sequences in the C1 regulatory domain of protein kinase C. The C-terminal half of n-chimaerin has 42% identity with the C-terminal region (amino acid residues 1050 to 1225) of BCR, the product of the breakpoint cluster region gene involved in Philadelphia (Ph') chromosome translocation. n-Chimaerin mRNA (2.2 x 10(3) base-pairs) is specifically expressed in the brain, with the highest amounts being in the hippocampus and cerebral cortex. The mRNA has a neuronal distribution and is expressed in neuroblastoma cells, but not in C6 glioma or primary astrocyte cultures. The similarity of two separate regions of n-chimaerin to domains of protein kinase C and BCR has intriguing implications with respect to its evolutionary origins, its function in the brain and potential phorbol-ester-binding properties.
- Ono Y et al.
- Phorbol ester binding to protein kinase C requires a cysteine-rich zinc-finger-like sequence.
- Proc Natl Acad Sci U S A. 1989; 86: 4868-71
- Display abstract
Protein kinase C normally has a tandem repeat of a characteristic cysteine-rich sequence in C1, the conserved region of the regulatory domain. These sequences resemble the DNA-binding zinc finger domain. For the gamma subspecies of rat brain protein kinase C, various deletion and point mutants in this domain were constructed, and the mutated proteins were expressed in Escherichia coli by using the T7 expression system. Radioactive phorbol 12,13-dibutyrate binding analysis indicated that a cysteine-rich zinc-finger-like sequence was essential for protein kinase C to bind phorbol ester and that one of two sequences was sufficient for the phorbol ester binding. Conserved region C2, another region in the regulatory domain, was apparently needed for the enzyme to require Ca2+ for phorbol ester binding activity.
- Disease (disease genes where sequence variants are found in this domain)
-
SwissProt sequences and OMIM curated human diseases associated with missense mutations within the C1 domain.
Protein Disease Protein kinase C gamma type (P05129) (SMART) OMIM:176980: PROTEIN KINASE C, GAMMA; PRKCG - Metabolism (metabolic pathways involving proteins which contain this domain)
-
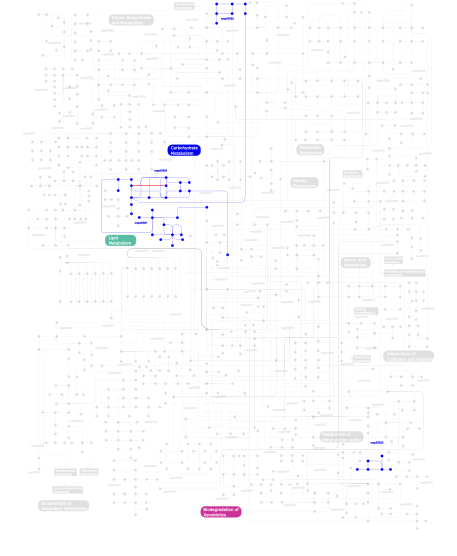
Click the image to view the interactive version of the map in iPath% proteins involved KEGG pathway ID Description 6.11 map04070 Phosphatidylinositol signaling system 5.53 map04510 Focal adhesion 4.88 map04530 Tight junction 4.73 map04670 Leukocyte transendothelial migration 4.73 map04010 MAPK signaling pathway 4.73 map04650 Natural killer cell mediated cytotoxicity 4.29 map05223 Non-small cell lung cancer 4.00 map04810 Regulation of actin cytoskeleton 4.00  map00564
map00564Glycerophospholipid metabolism 4.00  map00561
map00561Glycerolipid metabolism 3.49 map05214 Glioma 3.49 map04720 Long-term potentiation 3.49 map04012 ErbB signaling pathway 3.49 map04730 Long-term depression 3.28 map04310 Wnt signaling pathway 2.55 map04370 VEGF signaling pathway 2.55 map04916 Melanogenesis 2.55 map04540 Gap junction 2.40 map04910 Insulin signaling pathway 2.04 map05219 Bladder cancer 2.04 map04020 Calcium signaling pathway 1.75 map04664 Fc epsilon RI signaling pathway 1.46 map05213 Endometrial cancer 1.46 map05212 Pancreatic cancer 1.46 map05220 Chronic myeloid leukemia 1.46 map05210 Colorectal cancer 1.46 map05215 Prostate cancer 1.46 map05211 Renal cell carcinoma 1.46 map05221 Acute myeloid leukemia 1.46 map05218 Melanoma 1.46 map04914 Progesterone-mediated oocyte maturation 1.24 map04662 B cell receptor signaling pathway 1.24 map04350 TGF-beta signaling pathway 1.24 map04360 Axon guidance 1.24 map04660 T cell receptor signaling pathway 0.51 map05216 Thyroid cancer 0.51 map04912 GnRH signaling pathway 0.51 map04150 mTOR signaling pathway 0.15  map00562
map00562Inositol phosphate metabolism 0.07  map00632
map00632Benzoate degradation via CoA ligation 0.07 map04320 Dorso-ventral axis formation This information is based on mapping of SMART genomic protein database to KEGG orthologous groups. Percentage points are related to the number of proteins with C1 domain which could be assigned to a KEGG orthologous group, and not all proteins containing C1 domain. Please note that proteins can be included in multiple pathways, ie. the numbers above will not always add up to 100%.
- Structure (3D structures containing this domain)
3D Structures of C1 domains in PDB
PDB code Main view Title 1faq 
RAF-1 CYSTEINE RICH DOMAIN, NMR, 27 STRUCTURES 1far 
RAF-1 CYSTEINE RICH DOMAIN, NMR, MINIMIZED AVERAGE STRUCTURE 1kbe 
Solution structure of the cysteine-rich C1 domain of Kinase Suppressor of Ras 1kbf 
Solution Structure of the Cysteine-Rich C1 Domain of Kinase Suppressor of Ras 1ptq 
PROTEIN KINASE C DELTA CYS2 DOMAIN 1ptr 
PROTEIN KINASE C DELTA CYS2 DOMAIN COMPLEXED WITH PHORBOL-13-ACETATE 1r79 
Solution Structure of The C1 Domain of The Human Diacylglycerol Kinase Delta 1rfh 
Solution structure of the C1 domain of Nore1, a novel Ras effector 1tbn 
NMR STRUCTURE OF A PROTEIN KINASE C-G PHORBOL-BINDING DOMAIN, MINIMIZED AVERAGE STRUCTURE 1tbo 
NMR STRUCTURE OF A PROTEIN KINASE C-G PHORBOL-BINDING DOMAIN, 30 STRUCTURES 1xa6 
Crystal Structure of the Human Beta2-Chimaerin 1y8f 
Solution structure of the munc13-1 C1-domain 2db6 
Solution structure of RSGI RUH-051, a C1 domain of STAC3 from human cDNA 2e73 
Solution structure of the phorbol esters/diacylglycerol binding domain of protein kinase C gamma 2eli 
Solution structure of the second Phorbol esters/diacylglycerol binding domain of human Protein kinase C alpha type 2enn 
Solution structure of the first C1 domain from human protein kinase C theta 2enz 
Solution structure of the second C1 domain from human protein kinase C theta 2fnf 
C1 domain of Nore1 2ku3 
Solution structure of BRD1 PHD1 finger 2row 
The C1 domain of ROCK II 2vrw 
Critical structural role for the PH and C1 domains of the Vav1 exchange factor 2yuu 
Solution structure of the first Phorbol esters/diacylglycerol binding domain of human Protein kinase C, delta 3bji 
Structural Basis of Promiscuous Guanine Nucleotide Exchange by the T-Cell Essential Vav1 3cxl 
Crystal structure of human chimerin 1 (CHN1) 3ky9 
Autoinhibited Vav1 3pfq 
Crystal Structure and Allosteric Activation of Protein Kinase C beta II 3uej 
Structural and functional characterization of an anesthetic binding site in the second cysteine-rich domain of protein kinase Cdelta 3uey 
Structural and functional characterization of an anesthetic binding site in the second cysteine-rich domain of protein kinase Cdelta 3uff 
Structural and functional characterization of an anesthetic binding site in the second cysteine-rich domain of protein kinase Cdelta 3ugd 
Structural and functional characterization of an anesthetic binding site in the second cysteine-rich domain of protein kinase C delta 3ugi 
Structural and functional characterization of an anesthetic binding site in the second cysteine-rich domain of protein kinase C delta 3ugl 
Structural and functional characterization of an anesthetic binding site in the second cysteine-rich domain of protein kinase C delta 4b6d 
Structure of the atypical C1 domain of MgcRacGAP 4fkd 
Identification of the Activator Binding Residues in the Second Cysteine-Rich Regulatory Domain of Protein Kinase C Theta 4l9m 
Autoinhibited state of the Ras-specific exchange factor RasGRP1 - Links (links to other resources describing this domain)
-
PFAM DAG_PE-bind INTERPRO IPR002219

