The domain within your query sequence starts at position 105 and ends at position 216; the E-value for the CLECT domain shown below is 1.96e-18.
CPKEWILYSHNCYYIGMERKSWNDSLVSCISKNCSLLYIDSEEEQDFLQSLSLVSWTGIL RKGRGQPWVWKKDSTFKPKIAEILHDECNCAMMSASGLTADSCTTLHPYLCK
CLECTC-type lectin (CTL) or carbohydrate-recognition domain (CRD) |
|---|
| SMART accession number: | SM00034 |
|---|---|
| Description: | Many of these domains function as calcium-dependent carbohydrate binding modules. |
| Interpro abstract (IPR001304): | A number of different families of proteins share a conserved domain which was first characterised in some animal lectins and which seem to function as a calcium-dependent carbohydrate-recognition domain [ (PUBMED:3290208) (PUBMED:8341801) ]. This domain, which is known as the C-type lectin domain (CTL) or as the carbohydrate-recognition domain (CRD), consists of about 110 to 130 residues. There are four cysteines which are perfectly conserved and involved in two disulphide bonds. There are proteins with modules similar in overall structure to CRDs that serve functions other than sugar binding. Therefore, a more general term C-type lectin-like domain was introduced to refer to such domains, although both terms C-type lectin and C-type lectin-like are sometimes used interchangeably [ (PUBMED:16336259) ]. C-type lectins can be further divided into seven subgroups based on additional non-lectin domains and gene structure: (I) hyalectans, (II) asialoglycoprotein receptors, (III) collectins, (IV) selectins, (V) NK group transmembrane receptors, (VI) macrophage mannose receptors, and (VII) simple (single domain) lectins [ (PUBMED:15476922) ]. Lectins are a diverse group of proteins, both in terms of structure and activity. Carbohydrate binding ability may have evolved independently and sporadically in numerous unrelated families, where each evolved a structure that was conserved to fulfil some other activity and function. In general, animal lectins act as recognition molecules within the immune system, their functions involving defence against pathogens, cell trafficking, immune regulation and the prevention of autoimmunity [ (PUBMED:14519388) ]. |
| Family alignment: |
There are 74207 CLECT domains in 53821 proteins in SMART's nrdb database.
Click on the following links for more information.
- Evolution (species in which this domain is found)
-
Taxonomic distribution of proteins containing CLECT domain.
This tree includes only several representative species. The complete taxonomic breakdown of all proteins with CLECT domain is also avaliable.
Click on the protein counts, or double click on taxonomic names to display all proteins containing CLECT domain in the selected taxonomic class.
- Literature (relevant references for this domain)
-
Primary literature is listed below; Automatically-derived, secondary literature is also avaliable.
- Elgavish S, Shaanan B
- Lectin-carbohydrate interactions: different folds, common recognition principles.
- Trends Biochem Sci. 1997; 22: 462-7
- Display abstract
Lectins can be found in many organisms and are involved in a multitude of cellular processes that depend on specific recognition of complex carbohydrates. The stereochemical principles underlying the recognition process have been the subject of extensive biochemical and structural studies. When examined from the viewpoint of the bound sugar, the structural information accumulated so far on lectins and other proteins that are specific to galactose and glucose (or mannose), provides suggestive evidence for distinct ligand-dependent distribution of hydrogen-bond partners in the combining site.
- Weis WI, Drickamer K
- Structural basis of lectin-carbohydrate recognition.
- Annu Rev Biochem. 1996; 65: 441-73
- Display abstract
Lectins are responsible for cell surface sugar recognition in bacteria, animals, and plants. Examples include bacterial toxins; animal receptors that mediate cell-cell interactions, uptake of glycoconjugates, and pathogen neutralization; and plant toxins and mitogens. The structural basis for selective sugar recognition by members of all of these groups has been investigated by x-ray crystallography. Mechanisms for sugar recognition have evolved independently in diverse protein structural frameworks, but share some key features. Relatively low affinity binding sites for monosaccharides are formed at shallow indentations on protein surfaces. Selectivity is achieved through a combination of hydrogen bonding to the sugar hydroxyl groups with van der Waals packing, often including packing of a hydrophobic sugar face against aromatic amino acid side chains. Higher selectivity of binding is achieved by extending binding sites through additional direct and water-mediated contacts between oligosaccharides and the protein surface. Dramatically increased affinity for oligosaccharides results from clustering of simple binding sites in oligomers of the lectin polypeptides. The geometry of such oligomers helps to establish the ability of the lectins to distinguish surface arrays of polysaccharides in some instances and to crosslink glycoconjugates in others.
- Holmskov U, Malhotra R, Sim RB, Jensenius JC
- Collectins: collagenous C-type lectins of the innate immune defense system.
- Immunol Today. 1994; 15: 67-74
- Display abstract
Collectins are humoral lectins found in mammals and birds. They are oligomers whose subunits comprise three polypeptide chains each containing a collagenous section and a C-terminal lectin domain. They are related structurally and functionally to the first component of the classical complement pathway, C1q, and seem to serve important roles in innate immunity through opsonization and complement activation. The lectin domains bind carbohydrates on microorganisms, while the collagenous regions are ligands for the collectin receptor on phagocytes and also mediate C1q-independent activation of the classical complement pathway.
- Drickamer K
- Two distinct classes of carbohydrate-recognition domains in animal lectins.
- J Biol Chem. 1988; 263: 9557-60
- Metabolism (metabolic pathways involving proteins which contain this domain)
-
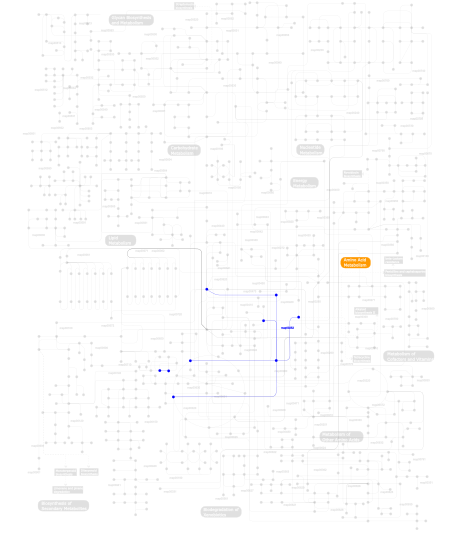
Click the image to view the interactive version of the map in iPath% proteins involved KEGG pathway ID Description 25.00 map04650 Natural killer cell mediated cytotoxicity 20.69 map04514 Cell adhesion molecules (CAMs) 19.83 map04610 Complement and coagulation cascades 18.97 map04612 Antigen processing and presentation 6.03 map03320 PPAR signaling pathway 5.17 map04662 B cell receptor signaling pathway 3.45 map04640 Hematopoietic cell lineage 0.86  map00252
map00252Alanine and aspartate metabolism This information is based on mapping of SMART genomic protein database to KEGG orthologous groups. Percentage points are related to the number of proteins with CLECT domain which could be assigned to a KEGG orthologous group, and not all proteins containing CLECT domain. Please note that proteins can be included in multiple pathways, ie. the numbers above will not always add up to 100%.
- Structure (3D structures containing this domain)
3D Structures of CLECT domains in PDB
PDB code Main view Title 1afa 
STRUCTURAL BASIS OF GALACTOSE RECOGNITION IN C-TYPE ANIMAL LECTINS 1afb 
STRUCTURAL BASIS OF GALACTOSE RECOGNITION IN C-TYPE ANIMAL LECTINS 1afd 
STRUCTURAL BASIS OF GALACTOSE RECOGNITION IN C-TYPE ANIMAL LECTINS 1b08 
LUNG SURFACTANT PROTEIN D (SP-D) (FRAGMENT) 1b6e 
HUMAN CD94 1bch 
MANNOSE-BINDING PROTEIN-A MUTANT (QPDWGH) COMPLEXED WITH N-ACETYL-D-GALACTOSAMINE 1bcj 
MANNOSE-BINDING PROTEIN-A MUTANT (QPDWGHV) COMPLEXED WITH N-ACETYL-D-GALACTOSAMINE 1bj3 
CRYSTAL STRUCTURE OF COAGULATION FACTOR IX-BINDING PROTEIN (IX-BP) FROM VENOM OF HABU SNAKE WITH A HETERODIMER OF C-TYPE LECTIN DOMAINS 1buu 
ONE HO3+ FORM OF RAT MANNOSE-BINDING PROTEIN A 1bv4 
APO-MANNOSE-BINDING PROTEIN-C 1byf 
STRUCTURE OF TC14; A C-TYPE LECTIN FROM THE TUNICATE POLYANDROCARPA MISAKIENSIS 1c3a 
CRYSTAL STRUCTURE OF FLAVOCETIN-A FROM THE HABU SNAKE VENOM, A NOVEL CYCLIC TETRAMER OF C-TYPE LECTIN-LIKE HETERODIMERS 1dv8 
CRYSTAL STRUCTURE OF THE CARBOHYDRATE RECOGNITION DOMAIN OF THE H1 SUBUNIT OF THE ASIALOGLYCOPROTEIN RECEPTOR 1e87 
Human CD69 - trigonal form 1e8i 
HUMAN CD69 - TETRAGONAL FORM 1egg 
STRUCTURE OF A C-TYPE CARBOHYDRATE-RECOGNITION DOMAIN (CRD-4) FROM THE MACROPHAGE MANNOSE RECEPTOR 1egi 
STRUCTURE OF A C-TYPE CARBOHYDRATE-RECOGNITION DOMAIN (CRD-4) FROM THE MACROPHAGE MANNOSE RECEPTOR 1esl 
INSIGHT INTO E-SELECTIN(SLASH)LIGAND INTERACTION FROM THE CRYSTAL STRUCTURE AND MUTAGENESIS OF THE LEC(SLASH)EGF DOMAINS 1fif 
N-ACETYLGALACTOSAMINE-SELECTIVE MUTANT OF MANNOSE-BINDING PROTEIN-A (QPDWG-HDRPY) 1fih 
N-ACETYLGALACTOSAMINE BINDING MUTANT OF MANNOSE-BINDING PROTEIN A (QPDWG-HDRPY), COMPLEX WITH N-ACETYLGALACTOSAMINE 1fm5 
CRYSTAL STRUCTURE OF HUMAN CD69 1fvu 
CRYSTAL STRUCTURE OF BOTROCETIN 1g1q 
Crystal structure of P-selectin lectin/EGF domains 1g1r 
Crystal structure of P-selectin lectin/EGF domains complexed with SLeX 1g1s 
P-SELECTIN LECTIN/EGF DOMAINS COMPLEXED WITH PSGL-1 PEPTIDE 1g1t 
CRYSTAL STRUCTURE OF E-SELECTIN LECTIN/EGF DOMAINS COMPLEXED WITH SLEX 1gz2 
Crystal structure of the Ovocleidin-17 a major protein of the Gallus gallus eggshell calcified layer. 1h8u 
Crystal Structure of the Eosinophil Major Basic Protein at 1.8Ã…: An Atypical Lectin with a Paradigm Shift in Specificity 1hq8 
CRYSTAL STRUCTURE OF THE MURINE NK CELL-ACTIVATING RECEPTOR NKG2D AT 1.95 A 1htn 
HUMAN TETRANECTIN, A TRIMERIC PLASMINOGEN BINDING PROTEIN WITH AN ALPHA-HELICAL COILED COIL 1hup 
HUMAN MANNOSE BINDING PROTEIN CARBOHYDRATE RECOGNITION DOMAIN TRIMERIZES THROUGH A TRIPLE ALPHA-HELICAL COILED-COIL 1hyr 
CRYSTAL STRUCTURE OF HUMAN MICA IN COMPLEX WITH NATURAL KILLER CELL RECEPTOR NKG2D 1ijk 
The von Willebrand Factor mutant (I546V) A1 domain-botrocetin Complex 1iod 
CRYSTAL STRUCTURE OF THE COMPLEX BETWEEN THE COAGULATION FACTOR X BINDING PROTEIN FROM SNAKE VENOM AND THE GLA DOMAIN OF FACTOR X 1ixx 
CRYSTAL STRUCTURE OF COAGULATION FACTORS IX/X-BINDING PROTEIN (IX/X-BP) FROM VENOM OF HABU SNAKE WITH A HETERODIMER OF C-TYPE LECTIN DOMAINS 1j34 
Crystal Structure of Mg(II)-and Ca(II)-bound Gla Domain of Factor IX Complexed with Binding Protein 1j35 
Crystal Structure of Ca(II)-bound Gla Domain of Factor IX Complexed with Binding Protein 1ja3 
Crystal Structure of the Murine NK Cell Inhibitory Receptor Ly-49I 1jwi 
Crystal Structure of Bitiscetin, a von Willeband Factor-dependent Platelet Aggregation Inducer. 1jzn 
crystal structure of a galactose-specific C-type lectin 1k9i 
Complex of DC-SIGN and GlcNAc2Man3 1k9j 
Complex of DC-SIGNR and GlcNAc2Man3 1kcg 
NKG2D in complex with ULBP3 1kg0 
Structure of the Epstein-Barr Virus gp42 Protein Bound to the MHC class II Receptor HLA-DR1 1kmb 
SELECTIN-LIKE MUTANT OF MANNOSE-BINDING PROTEIN A 1kwt 
Rat mannose binding protein A (native, MPD) 1kwu 
Rat mannose binding protein A complexed with a-Me-Man 1kwv 
Rat mannose binding protein A complexed with a-Me-GlcNAc 1kww 
Rat mannose protein A complexed with a-Me-Fuc. 1kwx 
Rat mannose protein A complexed with b-Me-Fuc. 1kwy 
Rat mannose protein A complexed with man-a13-man. 1kwz 
Rat mannose protein A (H189V) complexed with Man-a13-Man 1kx0 
Rat mannose protein A (H189V I207V) complexed with man-a13-man 1kx1 
Rat mannose protein A complexed with Man6-GlcNAc2-Asn 1kza 
Complex of MBP-C and Man-a13-Man 1kzb 
Complex of MBP-C and trimannosyl core 1kzc 
Complex of MBP-C and high-affinity linear trimannose 1kzd 
Complex of MBP-C and GlcNAc-terminated core 1kze 
Complex of MBP-C and bivalent Man-terminated glycopeptide 1lit 
HUMAN LITHOSTATHINE 1mpu 
Crystal Structure of the free human NKG2D immunoreceptor 1msb 
STRUCTURE OF THE CALCIUM-DEPENDENT LECTIN DOMAIN FROM A RAT MANNOSE-BINDING PROTEIN DETERMINED BY MAD PHASING 1muq 
X-ray Crystal Structure of Rattlesnake Venom Complexed With Thiodigalactoside 1oz7 
Crystal structure of Echicetin from the venom of Indian saw-scaled viper (Echis carinatus) at 2.4 resolution 1p1z 
X-RAY CRYSTAL STRUCTURE OF THE LECTIN-LIKE NATURAL KILLER CELL RECEPTOR LY-49C BOUND TO ITS MHC CLASS I LIGAND H-2Kb 1p4l 
Crystal structure of NK receptor Ly49C mutant with its MHC class I ligand H-2Kb 1pw9 
High resolution crystal structure of an active recombinant fragment of human lung surfactant protein D 1pwb 
High resolution crystal structure of an active recombinant fragment of human lung surfactant protein D with maltose 1qdd 
CRYSTAL STRUCTURE OF HUMAN LITHOSTATHINE TO 1.3 A RESOLUTION 1qo3 
Complex between NK cell receptor Ly49A and its MHC class I ligand H-2Dd 1r13 
Carbohydrate recognition and neck domains of surfactant protein A (SP-A) 1r14 
Carbohydrate recognition and neck domains of surfactant protein A (Sp-A) containing samarium 1rdi 
MANNOSE-BINDING PROTEIN, SUBTILISIN DIGEST FRAGMENT COMPLEX WITH ALPHA-METHYL-L-FUCOPYRANOSIDE 1rdj 
MANNOSE-BINDING PROTEIN, SUBTILISIN DIGEST FRAGMENT COMPLEX WITH BETA-METHYL-L-FUCOPYRANOSIDE 1rdk 
MANNOSE-BINDING PROTEIN, SUBTILISIN DIGEST FRAGMENT COMPLEX WITH D-GALACTOSE 1rdl 
MANNOSE-BINDING PROTEIN, SUBTILISIN DIGEST FRAGMENT COMPLEX WITH ALPHA-METHYL-D-MANNOPYRANOSIDE (0.2 M) 1rdm 
MANNOSE-BINDING PROTEIN, SUBTILISIN DIGEST FRAGMENT COMPLEX WITH ALPHA-METHYL-D-MANNOPYRANOSIDE (1.3 M) 1rdn 
MANNOSE-BINDING PROTEIN, SUBTILISIN DIGEST FRAGMENT COMPLEX WITH ALPHA-METHYL-D-N-ACETYLGLUCOSAMINIDE 1rdo 
MANNOSE-BINDING PROTEIN, SUBTILISIN DIGEST FRAGMENT 1rjh 
Structure of the Calcium Free Form of the C-type Lectin-like Domain of Tetranectin 1rtm 
TRIMERIC STRUCTURE OF A C-TYPE MANNOSE-BINDING PROTEIN 1sb2 
High resolution Structure determination of rhodocetin 1sl4 
Crystal Structure of DC-SIGN carbohydrate recognition domain complexed with Man4 1sl5 
Crystal Structure of DC-SIGN carbohydrate recognition domain complexed with LNFP III (Dextra L504). 1sl6 
Crystal Structure of a fragment of DC-SIGNR (containg the carbohydrate recognition domain and two repeats of the neck) complexed with Lewis-x. 1t8c 
Structure of the C-type lectin domain of CD23 1t8d 
Structure of the C-type lectin domain of CD23 1tdq 
Structural basis for the interactions between tenascins and the C-type lectin domains from lecticans: evidence for a cross-linking role for tenascins 1tlg 
STRUCTURE OF A TUNICATE C-TYPE LECTIN COMPLEXED WITH D-GALACTOSE 1tn3 
THE C-TYPE LECTIN CARBOHYDRATE RECOGNITION DOMAIN OF HUMAN TETRANECTIN 1u0n 
The ternary von Willebrand Factor A1-glycoprotein Ibalpha-botrocetin complex 1u0o 
The mouse von Willebrand Factor A1-botrocetin complex 1uex 
Crystal structure of von Willebrand Factor A1 domain complexed with snake venom bitiscetin 1ukm 
Crystal structure of EMS16, an Antagonist of collagen receptor integrin alpha2beta1 (GPIa/IIa) 1umr 
Crystal structure of the platelet activator convulxin, a disulfide linked a4b4 cyclic tetramer from the venom of Crotalus durissus terrificus 1uos 
The Crystal Structure of the Snake Venom Toxin Convulxin 1uv0 
Pancreatitis-associated protein 1 from human 1v4l 
Crystal structure of a platelet agglutination factor isolated from the venom of Taiwan habu (Trimeresurus mucrosquamatus) 1v7p 
Structure of EMS16-alpha2-I domain complex 1wk1 
Solution structure of Lectin C-type domain derived from a hypothetical protein from C. elegans 1wmy 
Crystal Structure of C-type Lectin CEL-I from Cucumaria echinata 1wmz 
Crystal Structure of C-type Lectin CEL-I complexed with N-acetyl-D-galactosamine 1wt9 
crystal structure of Aa-X-bp-I, a snake venom protein with the activity of binding to coagulation factor X from Agkistrodon acutus 1x2t 
Crystal Structure of Habu IX-bp at pH 6.5 1x2w 
Crystal Structure of Apo-Habu IX-bp at pH 4.6 1xar 
Crystal Structure of a fragment of DC-SIGNR (containing the carbohydrate recognition domain and two repeats of the neck). 1xph 
Structure of DC-SIGNR and a portion of repeat domain 8 1y17 
crystal structure of Aa-X-bp-II, a snake venom protein with the activity of binding to coagulation factor X from Agkistrodon acutus 1ypo 
Human Oxidized Low Density Lipoprotein Receptor LOX-1 P3 1 21 Space Group 1ypq 
Human Oxidized Low Density Lipoprotein Receptor LOX-1 Dioxane Complex 1ypu 
Human Oxidized Low Density Lipoprotein Receptor LOX-1 C2 Space Group 1ytt 
YB SUBSTITUTED SUBTILISIN FRAGMENT OF MANNOSE BINDING PROTEIN-A (SUB-MBP-A), MAD STRUCTURE AT 110K 1yxj 
Crystal structure of human lectin-like oxidized low-density lipoprotein receptor 1 (LOX-1) at low pH 1yxk 
Crystal structure of human lectin-like oxidized low-density lipoprotein receptor 1 (LOX-1) disulfide-linked dimer 2afp 
THE SOLUTION STRUCTURE OF TYPE II ANTIFREEZE PROTEIN REVEALS A NEW MEMBER OF THE LECTIN FAMILY 2b6b 
Cryo EM structure of Dengue complexed with CRD of DC-SIGN 2bpd 
STRUCTURE OF MURINE DECTIN-1 2bpe 
STRUCTURE OF MURINE DECTIN-1 2bph 
STRUCTURE OF MURINE DECTIN-1 2brs 
EMBP Heparin complex 2c6u 
Crystal structure of human CLEC-2 (CLEC1B) 2cl8 
Dectin-1 in complex with beta-glucan 2e3x 
Crystal structure of Russell's viper venom metalloproteinase 2ggu 
crystal structure of the trimeric neck and carbohydrate recognition domain of human surfactant protein D in complex with maltotriose 2ggx 
Crystal structure of the trimer neck and carbohydrate recognition domain of human surfactant protein D in complex with p-nitrophenyl maltoside 2go0 
NMR solution structure of human pancreatitis-associated protein 2h2r 
Crystal structure of the human CD23 Lectin domain, apo form 2h2t 
CD23 Lectin domain, Calcium 2+-bound 2it5 
Crystal Structure of DCSIGN-CRD with man6 2it6 
Crystal Structure of DCSIGN-CRD with man2 2kmb 
COMPLEX OF 3'-NEUAC-LEWIS-X WITH A SELECTIN-LIKE MUTANT OF MANNOSE-BINDING PROTEIN A 2kv3 
Human Regenerating Gene Type IV (REG IV) PROTEIN, P91S mutant 2ls8 
Solution structure of human C-type lectin domain family 4 member D 2m94 
2M94 2msb 
STRUCTURE OF A C-TYPE MANNOSE-BINDING PROTEIN COMPLEXED WITH AN OLIGOSACCHARIDE 2mti 
2MTI 2orj 
crystal structure of the trimeric neck and carbohydrate recognition domain of human surfactant protein D in complex with N-acetyl mannosamine 2ork 
crystal structure of the trimeric neck and carbohydrate recognition domain of human surfactant protein D in complex with inositol-1-phosphate 2os9 
crystal structure of the trimeric neck and carbohydrate recognition domain of human surfactant protein D in complex with myoinositol 2ox8 
Human Scavenger Receptor C-type Lectin carbohydrate-recognition domain. 2ox9 
Mouse Scavenger Receptor C-type Lectin carbohydrate-recognition domain. 2py2 
Structure of Herring Type II Antifreeze Protein 2ria 
Crystal structure of the trimeric neck and carbohydrate recognition domain of human surfactant protein D in complex with D-glycero-D-manno-heptose 2rib 
Crystal structure of the trimeric neck and carbohydrate recognition domain of human surfactant protein D in complex with L-glycero-D-manno-heptose 2ric 
Crystal structure of the trimeric neck and carbohydrate recognition domain of human surfactant protein D in complex with L-glycero-D-manno-heptopyranosyl-(1-3)-L-glycero-D-manno-heptopyranose 2rid 
Crystal structure of the trimeric neck and carbohydrate recognition domain of human surfactant protein D in complex with Allyl 7-O-carbamoyl-L-glycero-D-manno-heptopyranoside 2rie 
Crystal structure of the trimeric neck and carbohydrate recognition domain of human surfactant protein D in complex with 2-deoxy-L-glycero-D-manno-heptose 2vrp 
Structure of rhodocytin 2vuv 
Crystal structure of Codakine at 1.3A resolution 2vuz 
Crystal structure of Codakine in complex with biantennary nonasaccharide at 1.7A resolution 2xr5 
Crystal structure of the complex of the carbohydrate recognition domain of human DC-SIGN with pseudo dimannoside mimic. 2xr6 
Crystal structure of the complex of the carbohydrate recognition domain of human DC-SIGN with pseudo trimannoside mimic. 2yhf 
1.9 Angstrom Crystal Structure of CLEC5A 2zib 
Crystal structure analysis of calcium-independent type II antifreeze protein 3als 
Crystal structure of CEL-IV 3alt 
Crystal structure of CEL-IV complexed with Melibiose 3alu 
Crystal structure of CEL-IV complexed with Raffinose 3bdw 
Human CD94/NKG2A 3bx4 
Crystal structure of the snake venom toxin aggretin 3c22 
Crystal structure of the carbohydrate recognition domain of human Langerin 3c8j 
The crystal structure of natural killer cell receptor Ly49C 3c8k 
The crystal structure of Ly49C bound to H-2Kb 3cad 
Crystal structure of Natural Killer Cell Receptor, Ly49G 3cck 
Human CD69 3cdg 
Human CD94/NKG2A in complex with HLA-E 3cfw 
L-selectin lectin and EGF domains 3cii 
Structure of NKG2A/CD94 bound to HLA-E 3dbz 
human surfactant protein D 3fd4 
Crystal Structure of Epstein-Barr virus gp42 protein 3ff7 
Structure of NK cell receptor KLRG1 bound to E-cadherin 3ff8 
Structure of NK cell receptor KLRG1 bound to E-cadherin 3ff9 
Structure of NK cell receptor KLRG1 3g81 
Crystal structure of the trimeric neck and carbohydrate recognition domain of human surfactant protein D in complex with alpha methyl mannoside 3g83 
Crystal structure of the trimeric neck and carbohydrate recognition domain of human surfactant protein D in complex with alpha 1,2 dimannose. 3g84 
Crystal structure of the trimeric neck and carbohydrate recognition domain of R343V mutant of human surfactant protein D in complex with alpha 1,2 dimannose. 3g8k 
Crystal structure of murine natural killer cell receptor, Ly49L4 3g8l 
Crystal structure of murine natural killer cell receptor, Ly49L4 3gpr 
Crystal structure of rhodocetin 3hup 
High-resolution structure of the extracellular domain of human CD69 3ikn 
Crystal structure of galactose bound trimeric human lung surfactant protein D 3ikp 
Crystal structure of inositol phosphate bound trimeric human lung surfactant protein D 3ikq 
Crystal structure of alpha 1-2 mannobiose bound trimeric human lung surfactant protein D 3ikr 
Crystal structure of alpha 1-4 mannobiose bound trimeric human lung surfactant protein D 3j82 
3J82 3kmb 
COMPLEX OF 3'-SULFO-LEWIS-X WITH A SELECTIN-LIKE MUTANT OF MANNOSE-BINDING PROTEIN A 3kqg 
Trimeric Structure of Langerin 3l9j 
Selection of a novel highly specific TNFalpha antagonist: Insight from the crystal structure of the antagonist-TNFalpha complex 3m9z 
Crystal Structure of extracellular domain of mouse NKR-P1A 3p5d 
Structure of the carbohydrate-recognition domain of human Langerin with man5 (Man alpha1-3(Man alpha1-6)Man alpha1-6)(Man- alpha1-3)Man 3p5e 
Structure of the carbohydrate-recognition domain of human Langerin with man4 (Man alpha1-3(Man alpha1-6)Man alpha1-6Man) 3p5f 
Structure of the carbohydrate-recognition domain of human Langerin with man2 (Man alpha1-2 Man) 3p5g 
Structure of the carbohydrate-recognition domain of human Langerin with Blood group B trisaccharide (Gal alpha1-3(Fuc alpha1-2)Gal) 3p5h 
Structure of the carbohydrate-recognition domain of human Langerin with Laminaritriose 3p5i 
Structure of the carbohydrate-recognition domain of human Langerin with 6-SO4-Gal-GlcNAc 3p7f 
Structure of the human Langerin carbohydrate recognition domain 3p7g 
Structure of the human Langerin carbohydrate recognition domain in complex with mannose 3p7h 
Structure of the human Langerin carbohydrate recognition domain in complex with maltose 3pak 
Crystal Structure of Rat Surfactant Protein A neck and carbohydrate recognition domain (NCRD) complexed with Mannose 3paq 
Surfactant Protein A neck and carbohydrate recognition domain (NCRD) complexed with alpha-methylmannose 3par 
Surfactant Protein-A neck and carbohydrate recognition domain (NCRD) in the absence of ligand 3pbf 
Surfactant Protein-A neck and carbohydrate recognition domain (NCRD) complexed with glycerol 3rs1 
Mouse C-type lectin-related protein Clrg 3t3a 
Crystal structure of H107R mutant of extracellular domain of mouse receptor NKR-P1A 3ubu 
Crystal structure of agkisacucetin, a GpIb-binding snaclec (snake C-type lectin) that inhibits platelet 3vlg 
Crystal structure of the W150A mutant LOX-1 CTLD showing impaired OxLDL binding 3vpp 
Crystal Structure of the Human CLEC9A C-type Lectin-Like Domain 3vyj 
Crystal structure of C-type lectin domain of murine dendritic cell inhibitory receptor 2 (apo form) 3vyk 
Crystal structure of C-type lectin domain of murine dendritic cell inhibitory receptor 2 in complex with N-glycan 3wbp 
Crystal structure of carbohydrate recognition domain of Blood Dendritic Cell Antigen-2 (BDCA2) lectin (crystal form-1) 3wbq 
Crystal structure of carbohydrate recognition domain of Blood Dendritic Cell Antigen-2 (BDCA2) lectin (crystal form-2) 3wbr 
Crystal structure of carbohydrate recognition domain of Blood Dendritic Cell Antigen-2 (BDCA2) lectin (crystal form-3) 3wh2 
Human Mincle in complex with citrate 3wh3 
human Mincle, ligand free form 3whd 
C-type lectin, human MCL 3wsr 
3WSR 3wwk 
3WWK 3zhg 
Crystallographic structure of the native mouse SIGN-R1 CRD domain 4ak8 
Structure of F241L mutant of langerin carbohydrate recognition domain. 4c16 
4C16 4c9f 
4C9F 4caj 
4CAJ 4csy 
4CSY 4dn8 
Structure of porcine surfactant protein D neck and carbohydrate recognition domain complexed with mannose 4e52 
Crystal structure of Haemophilus Eagan 4A polysaccharide bound human lung surfactant protein D 4ezm 
Crystal structure of the human IgE-Fc(epsilon)3-4 bound to its B cell receptor derCD23 4g96 
Crystal structure of calcium2+-free wild-type CD23 lectin domain 4g9a 
Crystal structure of calcium2+-bound wild-type CD23 lectin domain 4gi0 
Crystal structure of CD23 lectin domain mutant E249A 4gj0 
Crystal structure of CD23 lectin domain mutant S252A 4gjx 
Crystal structure of CD23 lectin domain mutant D258A 4gk1 
Crystal structure of CD23 lectin domain mutant D270A 4gko 
Crystal structure of the calcium2+-bound human IgE-Fc(epsilon)3-4 bound to its B cell receptor derCD23 4iop 
Crystal structure of NKp65 bound to its ligand KACL 4j6j 
Crystal structure of calcium2+-free wild-type CD23 lectin domain (crystal form A) 4j6k 
Crystal structure of calcium2+-free wild-type CD23 lectin domain (crystal form B) 4j6l 
Crystal structure of calcium2+-free wild-type CD23 lectin domain (crystal form C) 4j6m 
Crystal structure of calcium2+-free wild-type CD23 lectin domain (crystal form D) 4j6n 
Crystal structure of calcium2+-free wild-type CD23 lectin domain (crystal form E) 4j6p 
Crystal structure of calcium2+-free wild-type CD23 lectin domain (crystal form F) 4j6q 
Crystal structure of calcium2+-free wild-type CD23 lectin domain (crystal form G) 4jo8 
Crystal structure of the activating Ly49H receptor in complex with m157 (G1F strain) 4ki1 
Primitive triclinic crystal form of the human IgE-Fc(epsilon)3-4 bound to its B cell receptor derCD23 4kmb 
COMPLEX OF 4'-SULFO-LEWIS-X WITH A SELECTIN-LIKE MUTANT OF MANNOSE-BINDING PROTEIN A 4kzv 
Structure of the carbohydrate-recognition domain of the C-type lectin mincle bound to trehalose 4kzw 
Structure of the carbohydrate-recognition domain of the C-type lectin mincle 4m17 
Crystal Structure of Surfactant Protein-D D325A/R343V mutant 4m18 
Crystal Structure of Surfactant Protein-D D325A/R343V mutant in complex with trimannose (Man-a1,2Man-a1,2Man) 4mth 
Crystal structure of mature human RegIIIalpha 4n32 
Structure of langerin CRD with alpha-Me-GlcNAc. 4n33 
Structure of langerin CRD complexed with GlcNAc-beta1-3Gal-beta1-4Glc-beta-CH2CH2N3 4n34 
Structure of langerin CRD I313 with alpha-MeGlcNAc 4n35 
Structure of langerin CRD I313 complexed with GlcNAc-beta1-3Gal-beta1-4Glc-beta-CH2CH2N3 4n36 
Structure of langerin CRD I313 D288 complexed with Me-GlcNAc 4n37 
Structure of langerin CRD I313 D288 complexed with Me-Man 4n38 
Structure of langerin CRD I313 D288 complexed with GlcNAc-beta1-3Gal-beta1-4GlcNAc-beta-CH2CH2N3 4pdc 
Crystal structure of Cowpox virus CPXV018 (OMCP) bound to human NKG2D 4pp8 
Crystal structure of murine NK cell ligand RAE-1 beta in complex with NKG2D 4qkg 
4QKG 4qkh 
4QKH 4qki 
4QKI 4qkj 
4QKJ 4s0u 
4S0U 4uww 
4UWW 4uxm 
4UXM 4wco 
4WCO 4wqq 
4WQQ 4wr9 
4WR9 4wrc 
4WRC 4wre 
4WRE 4wrf 
4WRF 4wuw 
4WUW 4wux 
4WUX 4yli 
4YLI 4ymd 
4YMD 4zes 
4ZES 4zet 
4ZET 4zrv 
4ZRV 4zrw 
4ZRW 5ao5 
5AO5 5ao6 
5AO6 5b1r 
5B1R 5b1w 
5B1W 5b1x 
5B1X 5e4k 
5E4K 5e4l 
5E4L 5ew6 
5EW6 5ffr 
5FFR 5ffs 
5FFS 5fft 
5FFT 5j6g 
5J6G 5kth 
5KTH 5kti 
5KTI - Links (links to other resources describing this domain)
-
PFAM lectin_c INTERPRO IPR001304 PROSITE CLECT_DOMAIN

