The domain within your query sequence starts at position 1116 and ends at position 1149; the E-value for the EGF domain shown below is 1.62e-5.
EFSPPMVLPRTSPCDNFDCQNGAQCIIRINEPIC
EGFEpidermal growth factor-like domain. |
|---|
| SMART accession number: | SM00181 |
|---|---|
| Description: | - |
| Interpro abstract (IPR000742): | A sequence of about thirty to forty amino-acid residues long found in the sequence of epidermal growth factor (EGF) has been shown [ (PUBMED:2288911) (PUBMED:6334307) (PUBMED:6607417) (PUBMED:3282918) ] to be present, in a more or less conserved form, in a large number of other, mostly animal proteins. EGF is a polypeptide of about 50 amino acids with three internal disulfide bridges. It first binds with high affinity to specific cell-surface receptors and then induces their dimerization, which is essential for activating the tyrosine kinase in the receptor cytoplasmic domain, initiating a signal transduction that results in DNA synthesis and cell proliferation. A common feature of all EGF-like domains is that they are found in the extracellular domain of membrane-bound proteins or in proteins known to be secreted (exception: prostaglandin G/H synthase). The EGF-like domain includes six cysteine residues which have been shown to be involved in disulfide bonds. The structure of several EGF-like domains has been solved. The fold consists of two-stranded beta-sheet followed by a loop to a C-terminal short two-stranded sheet. |
| Family alignment: |
There are 539475 EGF domains in 145300 proteins in SMART's nrdb database.
Click on the following links for more information.
- Evolution (species in which this domain is found)
-
Taxonomic distribution of proteins containing EGF domain.
This tree includes only several representative species. The complete taxonomic breakdown of all proteins with EGF domain is also avaliable.
Click on the protein counts, or double click on taxonomic names to display all proteins containing EGF domain in the selected taxonomic class.
- Literature (relevant references for this domain)
-
Primary literature is listed below; Automatically-derived, secondary literature is also avaliable.
- Rao Z, Handford P, Mayhew M, Knott V, Brownlee GG, Stuart D
- The structure of a Ca(2+)-binding epidermal growth factor-like domain: its role in protein-protein interactions.
- Cell. 1995; 82: 131-41
- Display abstract
Various diverse extracellular proteins possess Ca(2+)-binding epidermal growth factor (EGF)-like domains, the function of which remains uncertain. We have determined, at high resolution (1.5 A), the crystal structure of such a domain, from human clotting factor IX, as a complex with Ca2+. The Ca2+ ligands form a classic pentagonal bipyramid with six ligands contributed by one polypeptide chain and the seventh supplied by a neighboring EGF-like domain. The crystal structure identifies the role of Ca2+ in maintaining the conformation of the N-terminal region of the domain, but more importantly demonstrates that Ca2+ can directly mediate protein-protein contacts. The observed crystal packing of the domains provides a plausible model for the association of multiple tandemly linked EGF-like domains in proteins such as fibrillin-1, Notch, and protein S. This model is consistent with the known functional data and suggests a general biological role for these domains.
- Holmes WE et al.
- Identification of heregulin, a specific activator of p185erbB2.
- Science. 1992; 256: 1205-10
- Display abstract
The proto-oncogene designated erbB2 or HER2 encodes a 185-kilodalton transmembrane tyrosine kinase (p185erbB2), whose overexpression has been correlated with a poor prognosis in several human malignancies. A 45-kilodalton protein heregulin-alpha (HRG-alpha) that specifically induced phosphorylation of p185erbB2 was purified from the conditioned medium of a human breast tumor cell line. Several complementary DNA clones encoding related HRGs were identified, all of which are similar to proteins in the epidermal growth factor family. Scatchard analysis of the binding of recombinant HRG to a breast tumor cell line expressing p185erbB2 showed a single high affinity binding site [dissociation constant (Kd) = 105 +/- 15 picomolar]. Heregulin transcripts were identified in several normal tissues and cancer cell lines. The HRGs may represent the natural ligands for p185erbB2.
- Engel J
- EGF-like domains in extracellular matrix proteins: localized signals for growth and differentiation?
- FEBS Lett. 1989; 251: 1-7
- Display abstract
Multidomain proteins of the extracellular matrix (ECM) play an important role in development and maintenance of cellular organization and in tissue repair. Several ECM proteins such as laminin, tenascin and thrombospondin contain domains with homology to epidermal growth factor (EGF) and exhibit growth promoting activity. The mitogenic activity of laminin is restricted to a fragment which consists of about 25 repeating domains with partial homology to EGF and comprises the rod-like inner regions of the three short arms of the four armed molecule. The mitogenic activity does not correlate with promotion of cell attachment and neurite outgrowth for which major functional sites have been found in other regions of the laminin molecule. It is suggested that EGF-like domains in laminin, in other ECM proteins and in the extracellular portions of some membrane proteins are signals for cellular growth and differentiation. Because they are integral parts of large molecules and often of supramolecular assemblies these domains are well suited to stimulate neighboring cells in a specific and vectorial way. This concept of localized growth or differentiation signals offers an attractive mechanism for the regulation of cellular development.
- Appella E, Weber IT, Blasi F
- Structure and function of epidermal growth factor-like regions in proteins.
- FEBS Lett. 1988; 231: 1-4
- Doolittle RF, Feng DF, Johnson MS
- Computer-based characterization of epidermal growth factor precursor.
- Nature. 1984; 307: 558-60
- Display abstract
The cDNA sequence of the precursor of mouse epidermal growth factor (EGFP) has recently been reported by two groups, both of whom noted the presence of repeated similar segments, each about 40 residues long. One of these repeat units overlaps with the sequence of epidermal growth factor itself. The sequence of epidermal growth factor has been reported to be similar to that of pancreatic secretory trypsin inhibitor (PSTI) and a somewhat better match has been found with part of the sequence of bovine factor X, one of the blood coagulating factors. We report here that there is an even stronger similarity between the sequences of some of the repeat units of epidermal growth factor precursor and certain segments in factor X. This sequence similarity is also apparent in comparisons with other blood coagulation factors. On the basis of these sequence comparisons we suggest a scheme for the evolution of the epidermal growth factor precursor. We have also identified certain structural features in the precursor sequence that bear on the way in which the mature epidermal growth factor is generated.
- Disease (disease genes where sequence variants are found in this domain)
-
SwissProt sequences and OMIM curated human diseases associated with missense mutations within the EGF domain.
Protein Disease EGF-containing fibulin-like extracellular matrix protein 1 (Q12805) (SMART) OMIM:601548: Doyne honeycomb degeneration of retina
OMIM:126600:
OMIM:126600: Doyne honeycomb retinal dystrophyVitamin K-dependent protein C (P04070) (SMART) OMIM:176860: Thrombophilia due to protein C deficiency ; Purpura fulminans, neonatal Thyroid peroxidase (P07202) (SMART) OMIM:274500: Thyroid iodine peroxidase deficiency ; Goiter, congenital ; Hypothyroidism, congenital Low-density lipoprotein receptor (P01130) (SMART) OMIM:143890: Hypercholesterolemia, familial Coagulation factor IX (P00740) (SMART) OMIM:306900: Hemophilia B ; Warfarin sensitivity Fibrillin-1 (P35555) (SMART) OMIM:134797: Marfan syndrome
OMIM:154700: Shprintzen-Goldberg syndrome
OMIM:182212: Ectopia lentis, familial ; MASS syndrome
OMIM:604308:Coagulation factor VII (P08709) (SMART) OMIM:227500: Factor VII deficiency ; {Myocardial infarction, decreased susceptibility to} Delta-like protein 3 (Q9NYJ7) (SMART) OMIM:602768: Spondylocostal dysostosis, autosomal recessive, 1
OMIM:277300:Fibrillin-2 (P35556) (SMART) OMIM:121050: Contractural arachnodactyly, congenital Vitamin K-dependent protein S (P07225) (SMART) OMIM:176880: Protein S deficiency - Metabolism (metabolic pathways involving proteins which contain this domain)
-
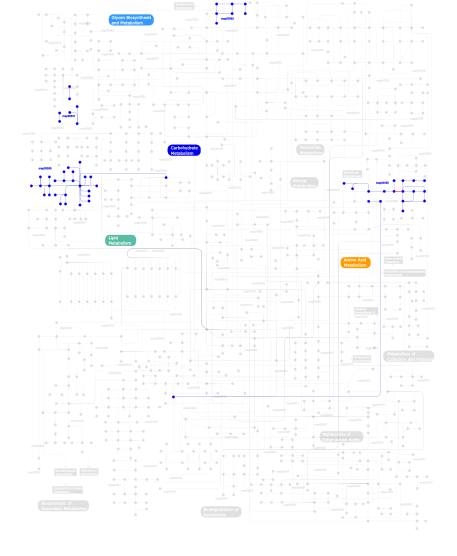
Click the image to view the interactive version of the map in iPath% proteins involved KEGG pathway ID Description 10.89 map04512 ECM-receptor interaction 10.51 map04510 Focal adhesion 9.08 map04610 Complement and coagulation cascades 9.08 map04330 Notch signaling pathway 8.17 map04012 ErbB signaling pathway 7.00 map04514 Cell adhesion molecules (CAMs) 4.02 map04350 TGF-beta signaling pathway 3.63 map04320 Dorso-ventral axis formation 3.37 map04810 Regulation of actin cytoskeleton 3.24 map04360 Axon guidance 2.85 map04310 Wnt signaling pathway 2.20 map04340 Hedgehog signaling pathway 2.08  map00590
map00590Arachidonic acid metabolism 1.95  map00531
map00531Glycosaminoglycan degradation 1.95 map01032 Glycan structures - degradation 1.82 map05212 Pancreatic cancer 1.82 map05214 Glioma 1.82 map05223 Non-small cell lung cancer 1.82 map05215 Prostate cancer 1.30 map04060 Cytokine-cytokine receptor interaction 1.17 map05217 Basal cell carcinoma 1.04 map05219 Bladder cancer 1.04 map05120 Epithelial cell signaling in Helicobacter pylori infection 1.04 map05213 Endometrial cancer 1.04 map04010 MAPK signaling pathway 1.04 map04912 GnRH signaling pathway 1.04 map04540 Gap junction 1.04 map05218 Melanoma 0.78 map05010 Alzheimer's disease 0.78 map05211 Renal cell carcinoma 0.52 map05222 Small cell lung cancer 0.26 map04670 Leukocyte transendothelial migration 0.13  map00350
map00350Tyrosine metabolism 0.13 map04140 Regulation of autophagy 0.13 map04640 Hematopoietic cell lineage 0.13  map00562
map00562Inositol phosphate metabolism 0.13 map04070 Phosphatidylinositol signaling system This information is based on mapping of SMART genomic protein database to KEGG orthologous groups. Percentage points are related to the number of proteins with EGF domain which could be assigned to a KEGG orthologous group, and not all proteins containing EGF domain. Please note that proteins can be included in multiple pathways, ie. the numbers above will not always add up to 100%.
- Structure (3D structures containing this domain)
3D Structures of EGF domains in PDB
PDB code Main view Title 1a3p 
ROLE OF THE 6-20 DISULFIDE BRIDGE IN THE STRUCTURE AND ACTIVITY OF EPIDERMAL GROWTH FACTOR, NMR, 20 STRUCTURES 1adx 
FIFTH EGF-LIKE DOMAIN OF THROMBOMODULIN (TMEGF5), NMR, 14 STRUCTURES 1cqe 
PROSTAGLANDIN H2 SYNTHASE-1 COMPLEX WITH FLURBIPROFEN 1cvu 
CRYSTAL STRUCTURE OF ARACHIDONIC ACID BOUND TO THE CYCLOOXYGENASE ACTIVE SITE OF COX-2 1cvw 
Crystal structure of active site-inhibited human coagulation factor VIIA (DES-GLA) 1cx2 
CYCLOOXYGENASE-2 (PROSTAGLANDIN SYNTHASE-2) COMPLEXED WITH A SELECTIVE INHIBITOR, SC-558 1ddx 
CRYSTAL STRUCTURE OF A MIXTURE OF ARACHIDONIC ACID AND PROSTAGLANDIN BOUND TO THE CYCLOOXYGENASE ACTIVE SITE OF COX-2: PROSTAGLANDIN STRUCTURE 1diy 
CRYSTAL STRUCTURE OF ARACHIDONIC ACID BOUND IN THE CYCLOOXYGENASE ACTIVE SITE OF PGHS-1 1dqb 
NMR STRUCTURE OF THROMBOMODULIN EGF(4-5) 1dx5 
Crystal structure of the thrombin-thrombomodulin complex 1ebv 
OVINE PGHS-1 COMPLEXED WITH SALICYL HYDROXAMIC ACID 1egf 
SOLUTION STRUCTURE OF MURINE EPIDERMAL GROWTH FACTOR DETERMINED BY NMR SPECTROSCOPY AND REFINED BY ENERGY MINIMIZATION WITH RESTRAINTS 1epg 
THREE-DIMENSIONAL NUCLEAR MAGNETIC RESONANCE STRUCTURES OF MOUSE EPIDERMAL GROWTH FACTOR IN ACIDIC AND PHYSIOLOGICAL PH SOLUTIONS 1eph 
THREE-DIMENSIONAL NUCLEAR MAGNETIC RESONANCE STRUCTURES OF MOUSE EPIDERMAL GROWTH FACTOR IN ACIDIC AND PHYSIOLOGICAL PH SOLUTIONS 1epi 
THREE-DIMENSIONAL NUCLEAR MAGNETIC RESONANCE STRUCTURES OF MOUSE EPIDERMAL GROWTH FACTOR IN ACIDIC AND PHYSIOLOGICAL PH SOLUTIONS 1epj 
THREE-DIMENSIONAL NUCLEAR MAGNETIC RESONANCE STRUCTURES OF MOUSE EPIDERMAL GROWTH FACTOR IN ACIDIC AND PHYSIOLOGICAL PH SOLUTIONS 1eqg 
THE 2.6 ANGSTROM MODEL OF OVINE COX-1 COMPLEXED WITH IBUPROFEN 1eqh 
THE 2.7 ANGSTROM MODEL OF OVINE COX-1 COMPLEXED WITH FLURBIPROFEN 1esl 
INSIGHT INTO E-SELECTIN(SLASH)LIGAND INTERACTION FROM THE CRYSTAL STRUCTURE AND MUTAGENESIS OF THE LEC(SLASH)EGF DOMAINS 1fe2 
CRYSTAL STRUCTURE OF DIHOMO-GAMMA-LINOLEIC ACID BOUND IN THE CYCLOOXYGENASE CHANNEL OF PROSTAGLANDIN ENDOPEROXIDE H SYNTHASE-1. 1fjs 
CRYSTAL STRUCTURE OF THE INHIBITOR ZK-807834 (CI-1031) COMPLEXED WITH FACTOR XA 1fsb 
STRUCTURE OF THE EGF DOMAIN OF P-SELECTIN, NMR, 19 STRUCTURES 1g1q 
Crystal structure of P-selectin lectin/EGF domains 1g1r 
Crystal structure of P-selectin lectin/EGF domains complexed with SLeX 1g1s 
P-SELECTIN LECTIN/EGF DOMAINS COMPLEXED WITH PSGL-1 PEPTIDE 1g1t 
CRYSTAL STRUCTURE OF E-SELECTIN LECTIN/EGF DOMAINS COMPLEXED WITH SLEX 1gk5 
Solution Structure the mEGF/TGFalpha44-50 chimeric growth factor 1gl4 
Nidogen-1 G2/Perlecan IG3 Complex 1hae 
HEREGULIN-ALPHA EPIDERMAL GROWTH FACTOR-LIKE DOMAIN, NMR, 20 STRUCTURES 1haf 
HEREGULIN-ALPHA EPIDERMAL GROWTH FACTOR-LIKE DOMAIN, NMR, MINIMIZED AVERAGE STRUCTURE 1hcg 
STRUCTURE OF HUMAN DES(1-45) FACTOR XA AT 2.2 ANGSTROMS RESOLUTION 1hre 
SOLUTION STRUCTURE OF THE EPIDERMAL GROWTH FACTOR-LIKE DOMAIN OF HEREGULIN-ALPHA, A LIGAND FOR P180ERB4 1hrf 
SOLUTION STRUCTURE OF THE EPIDERMAL GROWTH FACTOR-LIKE DOMAIN OF HEREGULIN-ALPHA, A LIGAND FOR P180ERB4 1ht5 
THE 2.75 ANGSTROM RESOLUTION MODEL OF OVINE COX-1 COMPLEXED WITH METHYL ESTER FLURBIPROFEN 1ht8 
THE 2.7 ANGSTROM RESOLUTION MODEL OF OVINE COX-1 COMPLEXED WITH ALCLOFENAC 1igx 
Crystal Structure of Eicosapentanoic Acid Bound in the Cyclooxygenase Channel of Prostaglandin Endoperoxide H Synthase-1. 1igz 
Crystal Structure of Linoleic acid Bound in the Cyclooxygenase Channel of Prostaglandin Endoperoxide H Synthase-1. 1ijq 
Crystal Structure of the LDL Receptor YWTD-EGF Domain Pair 1iox 
NMR Structure of human Betacellulin-2 1ip0 
NMR STRUCTURE OF HUMAN BETACELLULIN-2 1ivo 
Crystal Structure of the Complex of Human Epidermal Growth Factor and Receptor Extracellular Domains. 1j9c 
Crystal Structure of tissue factor-factor VIIa complex 1jbu 
Coagulation Factor VII Zymogen (EGF2/Protease) in Complex with Inhibitory Exosite Peptide A-183 1jl9 
Crystal Structure of Human Epidermal Growth Factor 1k36 
NMR Structure of human Epiregulin 1k37 
NMR Structure of human Epiregulin 1kig 
BOVINE FACTOR XA 1kli 
Cofactor-and substrate-assisted activation of factor VIIa 1klj 
Crystal structure of uninhibited factor VIIa 1mox 
Crystal Structure of Human Epidermal Growth Factor Receptor (residues 1-501) in complex with TGF-alpha 1mq5 
Crystal Structure of 3-chloro-N-[4-chloro-2-[[(4-chlorophenyl)amino]carbonyl]phenyl]-4-[(4-methyl-1-piperazinyl)methyl]-2-thiophenecarboxamide Complexed with Human Factor Xa 1mq6 
Crystal Structure of 3-chloro-N-[4-chloro-2-[[(5-chloro-2-pyridinyl)amino]carbonyl]-6-methoxyphenyl]-4-[[(4,5-dihydro-2-oxazolyl)methylamino]methyl]-2-thiophenecarboxamide Complexed with Human Factor Xa 1nql 
Structure of the extracellular domain of human epidermal growth factor (EGF) receptor in an inactive (low pH) complex with EGF. 1p9j 
Solution structure and dynamics of the EGF/TGF-alpha chimera T1E 1pge 
PROSTAGLANDIN H2 SYNTHASE-1 COMPLEXED WITH P-(2'-IODO-5'-THENOYL)HYDROTROPIC ACID (IODOSUPROFEN) 1pgf 
PROSTAGLANDIN H2 SYNTHASE-1 COMPLEXED WITH 1-(4-IODOBENZOYL)-5-METHOXY-2-METHYLINDOLE-3-ACETIC ACID (IODOINDOMETHACIN), CIS MODEL 1pgg 
PROSTAGLANDIN H2 SYNTHASE-1 COMPLEXED WITH 1-(4-IODOBENZOYL)-5-METHOXY-2-METHYLINDOLE-3-ACETIC ACID (IODOINDOMETHACIN), TRANS MODEL 1prh 
THE X-RAY CRYSTAL STRUCTURE OF THE MEMBRANE PROTEIN PROSTAGLANDIN H2 SYNTHASE-1 1pth 
THE STRUCTURAL BASIS OF ASPIRIN ACTIVITY INFERRED FROM THE CRYSTAL STRUCTURE OF INACTIVATED PROSTAGLANDIN H2 SYNTHASE 1pxx 
CRYSTAL STRUCTURE OF DICLOFENAC BOUND TO THE CYCLOOXYGENASE ACTIVE SITE OF COX-2 1q4g 
2.0 Angstrom Crystal Structure of Ovine Prostaglandin H2 Synthase-1, in complex with alpha-methyl-4-biphenylacetic acid 1qfk 
STRUCTURE OF HUMAN FACTOR VIIA AND ITS IMPLICATIONS FOR THE TRIGGERING OF BLOOD COAGULATION 1rfn 
HUMAN COAGULATION FACTOR IXA IN COMPLEX WITH P-AMINO BENZAMIDINE 1tpg 
F1-G MODULE PAIR RESIDUES 1-91 (C83S) OF TISSUE-TYPE PLASMINOGEN ACTIVATOR (T-PA) (NMR, 298K, PH2.95, REPRESENTATIVE STRUCTURE) 1u67 
Crystal Structure of Arachidonic Acid Bound to a Mutant of Prostagladin H Synthase-1 that Forms Predominantly 11-HPETE. 1v3x 
Factor Xa in complex with the inhibitor 1-[6-methyl-4,5,6,7-tetrahydrothiazolo(5,4-c)pyridin-2-yl] carbonyl-2-carbamoyl-4-(6-chloronaphth-2-ylsulphonyl)piperazine 1w7x 
Factor7 - 413 complex 1w8b 
Factor7 - 413 complex 1xdt 
COMPLEX OF DIPHTHERIA TOXIN AND HEPARIN-BINDING EPIDERMAL GROWTH FACTOR 1xfe 
Solution structure of the LA7-EGFA pair from the LDL receptor 1ygc 
Short Factor VIIa with a small molecule inhibitor 1yo8 
Structure of the C-terminal domain of human thrombospondin-2 1yuf 
TYPE ALPHA TRANSFORMING GROWTH FACTOR, NMR, 16 MODELS WITHOUT ENERGY MINIMIZATION 1yug 
TYPE ALPHA TRANSFORMING GROWTH FACTOR, NMR, 15 MODELS AFTER ECEPP/3 ENERGY MINIMIZATION 1z1y 
Crystal structure of Methylated Pvs25, an ookinete protein from Plasmodium vivax 1z27 
Crystal structure of Native Pvs25, an ookinete protein from Plasmodium vivax. 1z3g 
Crystal structure of complex between Pvs25 and Fab fragment of malaria transmission blocking antibody 2A8 1z6e 
Factor XA in complex with the inhibitor 1-(3'-amino-1,2-benzisoxazol-5'-yl)-n-(4-(2'-((dimethylamino)methyl)-1h-imidazol-1-yl)-2-fluorophenyl)-3-(trifluoromethyl)-1h-pyrazole-5-carboxamide (razaxaban; DPC906; BMS-561389) 1zaq 
FOURTH EGF-LIKE DOMAIN OF THROMBOMODULIN, NMR, 12 STRUCTURES 2adx 
FIFTH EGF-LIKE DOMAIN OF THROMBOMODULIN (TMEGF5), NMR, MINIMIZED AVERAGE STRUCTURE 2ayl 
2.0 Angstrom Crystal Structure of Manganese Protoporphyrin IX-reconstituted Ovine Prostaglandin H2 Synthase-1 Complexed With Flurbiprofen 2bmg 
Crystal structure of factor Xa in complex with 50 2bok 
Factor Xa - cation 2bq6 
Crystal structure of factor Xa in complex with 21 2bq7 
Crystal structure of factor Xa in complex with 43 2bqw 
CRYSTAL STRUCTURE OF FACTOR XA IN COMPLEX WITH COMPOUND 45 2bz6 
Orally available Factor7a inhibitor 2d1j 
Factor Xa in complex with the inhibitor 2-[[4-[(5-chloroindol-2-yl)sulfonyl]piperazin-1-yl] carbonyl]thieno[3,2-b]pyridine n-oxide 2ddu 
Crystal structure of the third repeat domain of reelin 2e26 
Crystal structure of two repeat fragment of reelin 2ei6 
FACTOR XA IN COMPLEX WITH THE INHIBITOR (-)-cis-N1-[(5-Chloroindol-2-yl)carbonyl]-N2-[(5-methyl-4,5,6,7-tetrahydrothiazolo[5,4-c]pyridin-2-yl)carbonyl]-1,2-cyclohexanediamine 2ei7 
FACTOR XA IN COMPLEX WITH THE INHIBITOR trans-N1-[(5-Chloroindol-2-yl)carbonyl]-N2-[(5-methyl-4,5,6,7-tetrahydrothiazolo[5,4-c]pyridin-2-yl)carbonyl]-1,2-cyclohexanediamine 2ei8 
FACTOR XA IN COMPLEX WITH THE INHIBITOR (1S,2R,4S)-N1-[(5-chloroindol-2-yl)carbonyl]-4-(N,N-dimethylcarbamoyl)-N2-[(5-methyl-4,5,6,7-tetrahydrothiazolo[5,4-c]pyridin-2-yl)carbonyl]-1,2-cyclohexanediamine 2fzz 
Factor Xa in complex with the inhibitor 1-(3-amino-1,2-benzisoxazol-5-yl)-6-(2'-(((3r)-3-hydroxy-1-pyrrolidinyl)methyl)-4-biphenylyl)-3-(trifluoromethyl)-1,4,5,6-tetrahydro-7h-pyrazolo[3,4-c]pyridin-7-one 2g00 
Factor Xa in complex with the inhibitor 3-(6-(2'-((dimethylamino)methyl)-4-biphenylyl)-7-oxo-3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-c]pyridin-1-yl)benzamide 2gd4 
Crystal Structure of the Antithrombin-S195A Factor Xa-Pentasaccharide Complex 2gy5 
Tie2 Ligand-Binding Domain Crystal Structure 2gy7 
Angiopoietin-2/Tie2 Complex Crystal Structure 2i9a 
Crystal structure of the free aminoterminal fragment of urokinase type plasminogen activator (ATF) 2i9b 
Crystal structure of ATF-urokinase receptor complex 2jkh 
Factor Xa - cation inhibitor complex 2k2s 
structure of the MIC1-GLD/MIC6-EGF complex from Toxoplasma gondii 2k2t 
Epidermal growth Factor-like domain 2 from Toxoplasma gondii Microneme protein 6 2kv4 
EGF 2m74 
1H, 13C and 15N assignments of the four N-terminal domains of human fibrillin-1 2oye 
Indomethacin-(R)-alpha-ethyl-ethanolamide bound to Cyclooxygenase-1 2oyu 
Indomethacin-(S)-alpha-ethyl-ethanolamide bound to Cyclooxygenase-1 2p16 
Factor Xa in Complex with the Inhibitor APIXABAN (BMS-562247) AKA 1-(4-METHOXYPHENYL)-7-OXO-6-(4-(2-OXO-1-PIPERIDINYL)PHENYL)-4,5,6,7-TETRAHYDRO-1H-PYRAZOLO[3, 4-C]PYRIDINE-3-CARBOXAMIDE 2p3f 
Crystal structure of the factor Xa/NAP5 complex 2p3t 
Crystal structure of human factor XA complexed with 3-Chloro-4-(2-methylamino-imidazol-1-ylmethyl)-thiophene-2-carboxylic acid [4-chloro-2-(5-chloro-pyridin-2-ylcarbamoyl)-6-methoxy-phenyl]-amide 2p3u 
Crystal structure of human factor XA complexed with 3-chloro-N-(4-chloro-2-{[(5-chloropyridin-2-yl)amino]carbonyl}-6-methoxyphenyl)-4-[(1-methyl-1H-imidazol-2-yl)methyl]thiophene-2-carboxamide {Pfizer 320663} 2p93 
Factor xa in complex with the inhibitor 5-chloro-N-(2-(4-(2-oxopyridin-1(2H)-yl)benzamido)ethyl)thiophene-2-carboxamide 2p94 
Factor xa in complex with the inhibitor 3-chloro-N-((1R,2S)-2-(4-(2-oxopyridin-1(2H)-yl)benzamido)cyclohexyl)-1H-indole-6-carboxamide 2p95 
Factor xa in complex with the inhibitor 5-chloro-N-((1R,2S)-2-(4-(2-oxopyridin-1(2H)-YL)benzamido) cyclopentyl)thiophene-2-carboxamide 2pe4 
Structure of Human Hyaluronidase 1, a Hyaluronan Hydrolyzing Enzyme Involved in Tumor Growth and Angiogenesis 2phb 
An Orally Efficacious Factor Xa Inhibitor 2pr3 
Factor XA inhibitor 2puq 
Crystal structure of active site inhibited coagulation factor VIIA in complex with soluble tissue factor 2q1j 
The discovery of glycine and related amino acid-based factor xa inhibitors 2ra0 
X-ray Structure of FXa in complex with 7-fluoroindazole 2rhp 
The Thrombospondin-1 Polymorphism Asn700Ser Associated with Cornoary Artery Disease Causes Local and Long-Ranging Changes in Protein Structure 2rnl 
Solution structure of the EGF-like domain from human Amphiregulin 2tgf 
THE SOLUTION STRUCTURE OF HUMAN TRANSFORMING GROWTH FACTOR ALPHA 2vvc 
Aminopyrrolidine Factor Xa inhibitor 2vvu 
Aminopyrrolidine Factor Xa inhibitor 2vvv 
Aminopyrrolidine-related triazole Factor Xa inhibitor 2vwl 
Aminopyrrolidine Factor Xa inhibitor 2vwm 
Aminopyrrolidine Factor Xa inhibitor 2vwn 
Aminopyrrolidine Factor Xa inhibitor 2vwo 
Aminopyrrolidine Factor Xa inhibitor 2w26 
Fator Xa in complex with BAY59-7939 2w3i 
Crystal Structure of FXa in complex with 4,4-disubstituted pyrrolidine-1,2-dicarboxamide inhibitor 2 2w3k 
Crystal Structure of FXa in complex with 4,4-disubstituted pyrrolidine-1,2-dicarboxamide inhibitor 1 2wft 
Crystal structure of the human HIP ectodomain 2wfx 
Crystal structure of the complex between human hedgehog-interacting protein HIP and Sonic Hedgehog in the presence of calcium 2wg3 
Crystal structure of the complex between human hedgehog-interacting protein HIP and desert hedgehog without calcium 2wg4 
Crystal structure of the complex between human hedgehog-interacting protein HIP and sonic hedgehog without calcium 2wph 
factor IXa superactive triple mutant 2wpi 
factor IXa superactive double mutant 2wpj 
factor IXa superactive triple mutant, NaCl-soaked 2wpk 
factor IXa superactive triple mutant, ethylene glycol-soaked 2wpl 
factor IXa superactive triple mutant, EDTA-soaked 2wpm 
factor IXa superactive mutant, EGR-CMK inhibited 2xbv 
Factor Xa in complex with a pyrrolidine-3,4-dicarboxylic acid inhibitor 2xbw 
Factor Xa in complex with a pyrrolidine-3,4-dicarboxylic acid inhibitor 2xbx 
Factor Xa in complex with a pyrrolidine-3,4-dicarboxylic acid inhibitor 2xby 
Factor Xa in complex with a pyrrolidine-3,4-dicarboxylic acid inhibitor 2xc0 
Factor Xa in complex with a pyrrolidine-3,4-dicarboxylic acid inhibitor 2xc4 
Factor Xa in complex with a pyrrolidine-3,4-dicarboxylic acid inhibitor 2xc5 
Factor Xa in complex with a pyrrolidine-3,4-dicarboxylic acid inhibitor 2y5f 
FACTOR XA - CATION INHIBITOR COMPLEX 2y5g 
FACTOR XA - CATION INHIBITOR COMPLEX 2y5h 
FACTOR XA - CATION INHIBITOR COMPLEX 2ygo 
WIF domain-EGF-like domain 1 of human Wnt inhibitory factor 1 in complex with 1,2-dipalmitoylphosphatidylcholine 2ygp 
WIF domain-EGF-like domain 1 Met77Trp of human Wnt inhibitory factor 1 in complex with 1,2-dipalmitoylphosphatidylcholine 2ygq 
WIF domain-epidermal growth factor (EGF)-like domains 1-3 of human Wnt inhibitory factor 1 in complex with 1,2- dipalmitoylphosphatidylcholine 3a7q 
Structural basis for specific recognition of reelin by its receptors 3asi 
Alpha-Neurexin-1 ectodomain fragment; LNS5-EGF3-LNS6 3bt1 
Structure of urokinase receptor, urokinase and vitronectin complex 3bt2 
Structure of urokinase receptor, urokinase and vitronectin complex 3c9a 
High Resolution Crystal Structure of Argos bound to the EGF domain of Spitz 3ca7 
High Resolution Crystal Structure of the EGF domain of Spitz 3cen 
Factor XA in complex with the inhibitor N-(2-(((5-chloro-2-pyridinyl) amino)sulfonyl)phenyl)-4-(2-oxo-1(2H)-pyridinyl)benzamide 3cfw 
L-selectin lectin and EGF domains 3cs7 
FACTOR XA IN COMPLEX WITH THE INHIBITOR 1-(4-methoxyphenyl)-6-(4-(1-(pyrrolidin-1-ylmethyl)cyclopropyl)phenyl)-3-(trifluoromethyl)-5,6-dihydro-1H-pyrazolo[3,4-c]pyridin-7(4H)-one 3e50 
Crystal structure of human insulin degrading enzyme in complex with transforming growth factor-alpha 3egf 
SOLUTION STRUCTURE OF MURINE EPIDERMAL GROWTH FACTOR DETERMINED BY NMR SPECTROSCOPY AND REFINED BY ENERGY MINIMIZATION WITH RESTRAINTS 3f1s 
Crystal structure of Protein Z complexed with protein Z-dependent inhibitor 3f6u 
Crystal structure of human Activated Protein C (APC) complexed with PPACK 3fby 
The crystal structure of the signature domain of cartilage oligomeric matrix protein. 3ffg 
Factor XA in complex with the inhibitor (R)-6-(2'-((3- HYDROXYPYRROLIDIN-1-YL)METHYL)BIPHENYL-4-YL)-1-(3-(5-OXO-4,5-DIHYDRO-1H-1,2,4-TRIAZOL-3-YL)PHENYL)-3-(TRIFLUOROMETHYL)-5,6-DIHYDRO-1H-PYRAZOLO[3,4-C]PYRIDIN- 7(4H)-ONE 3g5c 
Structural and biochemical studies on the ectodomain of human ADAM22 3gis 
Crystal Structure of Na-free Thrombin in Complex with Thrombomodulin 3h5c 
X-Ray Structure of Protein Z-Protein Z Inhibitor Complex 3ho3 
Crystal structure of Hedgehog-interacting protein (HHIP) 3ho4 
Crystal structure of Hedgehog-interacting protein (HHIP) 3ho5 
Crystal structure of Hedgehog-interacting protein (HHIP) and Sonic hedgehog (SHH) complex 3hs5 
X-ray crystal structure of arachidonic acid bound to the cyclooxygenase channel of cyclooxygenase-2 3hs6 
X-ray crystal structure of eicosapentaenoic acid bound to the cyclooxygenase channel of cyclooxygenase-2 3hs7 
X-ray crystal structure of docosahexaenoic acid bound to the cyclooxygenase channel of cyclooxygenase-2 3iit 
Factor XA in complex with a cis-1,2-diaminocyclohexane derivative 3kcg 
Crystal structure of the antithrombin-factor IXa-pentasaccharide complex 3kk6 
Crystal Structure of Cyclooxygenase-1 in complex with celecoxib 3kl6 
Discovery of Tetrahydropyrimidin-2(1H)-one derivative TAK-442: A potent, selective and orally active factor Xa inhibitor 3kqb 
Factor xa in complex with the inhibitor n-(3-fluoro-2'- (methylsulfonyl)biphenyl-4-yl)-1-(3-(5-oxo-4,5-dihydro-1h- 1,2,4-triazol-3-yl)phenyl)-3-(trifluoromethyl)-1h- pyrazole-5-carboxamide 3kqc 
Factor xa in complex with the inhibitor 6-(2'- (methylsulfonyl)biphenyl-4-yl)-1-(3-(5-oxo-4,5-dihydro-1h- 1,2,4-triazol-3-yl)phenyl)-3-(trifluoromethyl)-5,6- dihydro-1h-pyrazolo[3,4-c]pyridin-7(4h)-one 3kqd 
Factor xa in complex with the inhibitor 1-(3-(5-oxo-4,5- dihydro-1h-1,2,4-triazol-3-yl)phenyl)-6-(2'-(pyrrolidin-1- ylmethyl)biphenyl-4-yl)-3-(trifluoromethyl)-5,6-dihydro- 1h-pyrazolo[3,4-c]pyridin-7(4h)-one 3kqe 
Factor xa in complex with the inhibitor 3-methyl-1-(3-(5- oxo-4,5-dihydro-1h-1,2,4-triazol-3-yl)phenyl)-6-(2'- (pyrrolidin-1-ylmethyl)biphenyl-4-yl)-5,6-dihydro-1h- pyrazolo[3,4-c]pyridin-7(4h)-one 3krk 
X-ray crystal structure of arachidonic acid bound in the cyclooxygenase channel of L531F murine COX-2 3lc3 
Benzothiophene Inhibitors of Factor IXa 3lc5 
Selective Benzothiophine Inhibitors of Factor IXa 3liw 
Factor XA in complex with (R)-2-(1-ADAMANTYLCARBAMOYLAMINO)-3-(3-CARBAMIDOYL-PHENYL)-N-PHENETHYL-PROPIONIC ACID AMIDE 3ln0 
Structure of compound 5c-S bound at the active site of COX-2 3ln1 
Structure of celecoxib bound at the COX-2 active site 3ltf 
Crystal Structure of the Drosophila Epidermal Growth Factor Receptor ectodomain in complex with Spitz 3ltg 
Crystal structure of the Drosophila Epidermal Growth Factor Receptor ectodomain complexed with a low affinity Spitz mutant 3m36 
Factor XA in complex with the inhibitor 1-[3-(aminomethyl)phenyl]-N-[3-fluoro-2'-(methylsulfonyl)biphenyl-4-yl]-3-(trifluoromethyl)-1H-pyrazole-5-carboxamide (DPC423) 3m37 
Factor XA in complex with the inhibitor 1-[2-(aminomethyl)phenyl]-N-(3-fluoro-2'-sulfamoylbiphenyl-4-yl)-3-(trifluoromethyl)-1H-pyrazole-5-carboxamide (DPC602) 3mdl 
X-ray crystal structure of 1-arachidonoyl glycerol bound to the cyclooxygenase channel of cyclooxygenase-2 3mqe 
Structure of SC-75416 bound at the COX-2 active site 3n8v 
Crystal Structure of Unoccupied Cyclooxygenase-1 3n8w 
Crystal Structure of R120Q/Native Cyclooxygenase-1 Heterodimer mutant in complex with Flurbiprofen 3n8x 
Crystal Structure of Cyclooxygenase-1 in Complex with Nimesulide 3n8y 
Structure of Aspirin Acetylated Cyclooxygenase-1 in Complex with Diclofenac 3n8z 
Crystal Structure of Cyclooxygenase-1 in Complex with Flurbiprofen 3njp 
The Extracellular and Transmembrane Domain Interfaces in Epidermal Growth Factor Receptor Signaling 3nt1 
High resolution structure of naproxen:COX-2 complex. 3ntb 
Structure of 6-methylthio naproxen analog bound to mCOX-2. 3ntg 
Crystal structure of COX-2 with selective compound 23d-(R) 3olt 
X-ray crystal structure of arachidonic acid bound to the cyclooxygenase channel of R513H murine COX-2 3olu 
X-ray crystal structure of 1-arachidonoyl glycerol bound to the cyclooxygenase channel of R513H murine COX-2 3pgh 
CYCLOOXYGENASE-2 (PROSTAGLANDIN SYNTHASE-2) COMPLEXED WITH A NON-SELECTIVE INHIBITOR, FLURBIPROFEN 3poy 
Crystal Structure of the alpha-Neurexin-1 ectodomain, LNS 2-6 3q3k 
Factor Xa in complex with a phenylenediamine derivative 3q7d 
Structure of (R)-naproxen bound to mCOX-2. 3qcw 
Structure of neurexin 1 alpha (domains LNS1-LNS6), no splice inserts 3qh0 
X-ray crystal structure of palmitic acid bound to the cyclooxygenase channel of cyclooxygenase-2 3qmo 
X-ray crystal structure of NS-398 bound to the cyclooxygenase channel of cyclooxygenase-2 3r05 
Structure of neurexin 1 alpha (domains LNS1-LNS6), with splice insert SS3 3rr3 
Structure of (R)-flurbiprofen bound to mCOX-2 3s2k 
Structural basis of Wnt signaling inhibition by Dickkopf binding to LRP5/6. 3s8v 
Crystal structure of LRP6-Dkk1 complex 3s8z 
Crystal structure of LRP6-E3E4 3s94 
Crystal structure of LRP6-E1E2 3sob 
The structure of the first YWTD beta propeller domain of LRP6 in complex with a FAB 3soq 
The structure of the first YWTD beta propeller domain of LRP6 in complex with a DKK1 peptide 3sov 
The structure of a beta propeller domain in complex with peptide S 3tgf 
THE SOLUTION STRUCTURE OF HUMAN TRANSFORMING GROWTH FACTOR ALPHA 3tk5 
Factor Xa in complex with D102-4380 3tk6 
factor Xa in complex with D46-5241 3tzi 
X-ray crystal structure of arachidonic acid bound in the cyclooxygenase channel of G533V murine COX-2 3u73 
Crystal structure of stabilized human uPAR mutant in complex with ATF 3u7u 
Crystal structure of extracellular region of human epidermal growth factor receptor 4 in complex with neuregulin-1 beta 3v64 
Crystal Structure of agrin and LRP4 4a0p 
Crystal structure of LRP6P3E3P4E4 4bdw 
The structure of the FnI-EGF tandem domain of coagulation factor XII in complex with Holmium 4bdx 
The structure of the FnI-EGF tandem domain of coagulation factor XII 4c16 
4C16 4cox 
CYCLOOXYGENASE-2 (PROSTAGLANDIN SYNTHASE-2) COMPLEXED WITH A NON-SELECTIVE INHIBITOR, INDOMETHACIN 4csy 
4CSY 4dg6 
Crystal structure of domains 1 and 2 of LRP6 4e1g 
X-ray crystal structure of alpha-linolenic acid bound to the cyclooxygenase channel of cyclooxygenase-2 4fm5 
X-ray structure of des-methylflurbiprofen bound to murine COX-2 4ish 
Structure of FACTOR VIIA in complex with the inhibitor BMS-593214 also known as 2'-[(6R,6AR,11BR)-2-CARBAMIMIDOYL-6,6A,7,11B-TETRAHYDRO-5H-INDENO[2,1-C]QUINOLIN-6-YL]-5'-HYDROXY-4'-METHOXYBIPHENYL-4-CARBOXYLIC ACID 4isi 
Structure of FACTOR VIIA in complex with the inhibitor (6S)-N-(4-CARBAMIMIDOYLBENZYL)-1-CHLORO-3-(CYCLOBUTYLAMINO)-8,8-DIETHYL-4-OXO-4,6,7,8-TETRAHYDROPYRROLO[1,2-A]PYRAZINE-6-CARBOXAMIDE 4jyu 
Structure of factor VIIA in complex with the inhibitor (2R)-2-[(1-AMINOISOQUINOLIN-6-YL)AMINO]-2-[3-ETHOXY-4-(PROPAN-2-YLOXY)PHENYL]-N-(PHENYLSULFONYL)ETHANAMIDE 4jyv 
Structure of factor VIIA in complex with the inhibitor (2R)-2-[3-ETHOXY-4-(PROPAN-2-YLOXY)PHENYL]-2-(ISOQUINOLIN-6-YLAMINO)-N-[(3-SULFAMOYLPHENYL)SULFONYL]ETHANAMIDE 4jzd 
Structure of factor VIIA in complex with the inhibitor 2-{2-[(4-carbamimidoylphenyl)carbamoyl]-6-methoxypyridin-3-yl}-5-{[(2S)-1-hydroxy-3,3-dimethylbutan-2-yl]carbamoyl}benzoic acid 4jze 
Structure of factor VIIA in complex with the inhibitor 2-{2-[(1-aminoisoquinolin-6-yl)carbamoyl]-6-methoxypyridin-3-yl}-5-{[(2S)-1-hydroxy-3,3-dimethylbutan-2-yl]carbamoyl}benzoic acid 4jzf 
Structure of factor VIIA in complex with the inhibitor 2-{2-[(3-carbamoylphenyl)carbamoyl]-6-methoxypyridin-3-yl}-5-{[(2S)-1-hydroxy-3,3-dimethylbutan-2-yl]carbamoyl}benzoic acid 4k0v 
Structural basis for angiopoietin-1 mediated signaling initiation 4k24 
Structure of anti-uPAR Fab ATN-658 in complex with uPAR 4m10 
Crystal Structure of Murine Cyclooxygenase-2 Complex with Isoxicam 4m11 
Crystal Structure of Murine Cyclooxygenase-2 Complex with Meloxicam 4na9 
Factor VIIa in complex with the inhibitor 3'-amino-5'-[(2s,4r)-6-carbamimidoyl-4-phenyl-1,2,3,4-tetrahydroquinolin-2-yl]biphenyl-2-carboxylic acid 4ng9 
Factor viia in complex with the inhibitor (2R)-2-[(1-aminoisoquinolin-6-yl)amino]-2-[3-ethoxy-4-(propan-2-yloxy)phenyl]-n-(3-sulfamoylbenzyl)ethanamide 4nga 
Factor viia in complex with the inhibitor (2R)-2-[(1-aminoisoquinolin-6-yl)amino]-2-[3-ethoxy-4-(propan-2-yloxy)phenyl]-N-[2-(propan-2-ylsulfonyl)benzyl]ethanamide 4o1z 
Crystal Structure of Ovine Cyclooxygenase-1 Complex with Meloxicam 4otj 
4OTJ 4oty 
Crystal structure of lumiracoxib bound to the apo-mouse-cyclooxygenase-2 4ph9 
4PH9 4rrw 
4RRW 4rrx 
4RRX 4rry 
4RRY 4rrz 
4RRZ 4rs0 
4RS0 4rut 
4RUT 4tgf 
SOLUTION STRUCTURES OF HUMAN TRANSFORMING GROWTH FACTOR ALPHA DERIVED FROM 1*H NMR DATA 4x8s 
4X8S 4x8t 
4X8T 4x8u 
4X8U 4x8v 
4X8V 4xl1 
4XL1 4xlw 
4XLW 4yt6 
4YT6 4yt7 
4YT7 4yzu 
4YZU 4z0k 
4Z0K 4z0l 
4Z0L 4z6a 
4Z6A 4z80 
4Z80 4z81 
4Z81 4zae 
4ZAE 4zxx 
4ZXX 4zxy 
4ZXY 5cox 
UNINHIBITED MOUSE CYCLOOXYGENASE-2 (PROSTAGLANDIN SYNTHASE-2) 5e8d 
5E8D 5egm 
5EGM 5f19 
5F19 5f1a 
5F1A 5fdq 
5FDQ 5fww 
5FWW 5i46 
5I46 5ikq 
5IKQ 5ikr 
5IKR 5ikt 
5IKT 5ikv 
5IKV 5jvy 
5JVY 5jvz 
5JVZ 5jw1 
5JW1 5k0h 
5K0H 5kir 
5KIR 5kn5 
5KN5 5l2y 
5L2Y 5l2z 
5L2Z 5l30 
5L30 6cox 
CYCLOOXYGENASE-2 (PROSTAGLANDIN SYNTHASE-2) COMPLEXED WITH A SELECTIVE INHIBITOR, SC-558 IN I222 SPACE GROUP - Links (links to other resources describing this domain)
-
INTERPRO IPR000742 PFAM EGF PROSITE EGF_1

