The domain within your query sequence starts at position 78 and ends at position 97; the E-value for the HhH1 domain shown below is 2.55e2.
EDLALVSGVGATKLEQVKFE
HhH1Helix-hairpin-helix DNA-binding motif class 1 |
|---|
| SMART accession number: | SM00278 |
|---|---|
| Description: | - |
| Interpro abstract (IPR003583): | The HhH motif is a stretch of approximately 20 amino acids that is present in prokaryotic and eukaryotic non-sequence-specific DNA binding proteins [ (PUBMED:7664751) (PUBMED:9973609) (PUBMED:9987128) ]. The HhH motif is similar to, but distinct from, the HtH motif. Both of these motifs have two helices connected by a short turn. In the HtH motif the second helix binds to DNA with the helix in the major groove. This allows the contact between specific base and residues throughout the protein. In the HhH motif the second helix does not protrude from the surface of the protein and therefore cannot lie in the major groove of the DNA. Crystallographic studies suggest that the interaction of the HhH domain with DNA is mediated by amino acids located in the strongly conserved loop (L-P-G-V) and at the N-terminal end of the second helix [ (PUBMED:7664751) ]. This interaction could involve the formation of hydrogen bonds between protein backbone nitrogens and DNA phosphate groups [ (PUBMED:8692686) ]. The structural difference between the HtH and HhH domains is reflected at the functional level: whereas the HtH domain, found primarily in gene regulatory proteins and binds DNA in a sequence specific manner, the HhH domain is rather found in proteins involved in enzymatic activities and binds DNA with no sequence specificity [ (PUBMED:8692686) ]. |
| GO process: | DNA repair (GO:0006281) |
| GO function: | DNA binding (GO:0003677) |
| Family alignment: |
There are 311019 HhH1 domains in 190339 proteins in SMART's nrdb database.
Click on the following links for more information.
- Evolution (species in which this domain is found)
-
Taxonomic distribution of proteins containing HhH1 domain.
This tree includes only several representative species. The complete taxonomic breakdown of all proteins with HhH1 domain is also avaliable.
Click on the protein counts, or double click on taxonomic names to display all proteins containing HhH1 domain in the selected taxonomic class.
- Metabolism (metabolic pathways involving proteins which contain this domain)
-
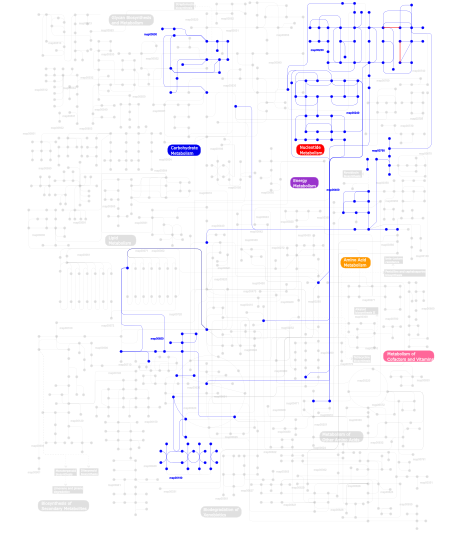
Click the image to view the interactive version of the map in iPath% proteins involved KEGG pathway ID Description 53.44 map03030 DNA replication 14.05  map00230
map00230Purine metabolism 13.96  map00240
map00240Pyrimidine metabolism 9.66 map03090 Type II secretion system 2.49 map03020 RNA polymerase 2.20 map05212 Pancreatic cancer 1.05  map00500
map00500Starch and sucrose metabolism 1.05  map00790
map00790Folate biosynthesis 0.86 map03010 Ribosome 0.48 map00970 Aminoacyl-tRNA biosynthesis 0.48  map00400
map00400Phenylalanine, tyrosine and tryptophan biosynthesis 0.10 map02060 Phosphotransferase system (PTS) 0.10  map00190
map00190Oxidative phosphorylation 0.10  map00650
map00650Butanoate metabolism This information is based on mapping of SMART genomic protein database to KEGG orthologous groups. Percentage points are related to the number of proteins with HhH1 domain which could be assigned to a KEGG orthologous group, and not all proteins containing HhH1 domain. Please note that proteins can be included in multiple pathways, ie. the numbers above will not always add up to 100%.
- Structure (3D structures containing this domain)
3D Structures of HhH1 domains in PDB
PDB code Main view Title 1b22 
RAD51 (N-TERMINAL DOMAIN) 1bdx 
E. COLI DNA HELICASE RUVA WITH BOUND DNA HOLLIDAY JUNCTION, ALPHA CARBONS AND PHOSPHATE ATOMS ONLY 1bgx 
TAQ POLYMERASE IN COMPLEX WITH TP7, AN INHIBITORY FAB 1bno 
NMR SOLUTION STRUCTURE OF THE N-TERMINAL DOMAIN OF DNA POLYMERASE BETA, MINIMIZED AVERAGE STRUCTURE 1bnp 
NMR SOLUTION STRUCTURE OF THE N-TERMINAL DOMAIN OF DNA POLYMERASE BETA, 55 STRUCTURES 1bpb 
CRYSTAL STRUCTURE OF RAT DNA POLYMERASE BETA: EVIDENCE FOR A COMMON POLYMERASE MECHANISM 1bpd 
CRYSTAL STRUCTURE OF RAT DNA POLYMERASE BETA: EVIDENCE FOR A COMMON POLYMERASE MECHANISM 1bpe 
CRYSTAL STRUCTURE OF RAT DNA POLYMERASE BETA; EVIDENCE FOR A COMMON POLYMERASE MECHANISM 1bpx 
DNA POLYMERASE BETA/DNA COMPLEX 1bpy 
HUMAN DNA POLYMERASE BETA COMPLEXED WITH GAPPED DNA AND DDCTP 1bpz 
HUMAN DNA POLYMERASE BETA COMPLEXED WITH NICKED DNA 1bvs 
RUVA COMPLEXED TO A HOLLIDAY JUNCTION. 1c7y 
E.COLI RUVA-HOLLIDAY JUNCTION COMPLEX 1cuk 
ESCHERICHIA COLI RUVA PROTEIN AT PH 4.9 AND ROOM TEMPERATURE 1d8l 
E. COLI HOLLIDAY JUNCTION BINDING PROTEIN RUVA NH2 REGION LACKING DOMAIN III 1dgs 
CRYSTAL STRUCTURE OF NAD+-DEPENDENT DNA LIGASE FROM T. FILIFORMIS 1dk2 
REFINED SOLUTION STRUCTURE OF THE N-TERMINAL DOMAIN OF DNA POLYMERASE BETA 1dk3 
REFINED SOLUTION STRUCTURE OF THE N-TERMINAL DOMAIN OF DNA POLYMERASE BETA 1doq 
THE C-TERMINAL DOMAIN OF THE RNA POLYMERASE ALPHA SUBUNIT FROM THERMUS THERMOPHILUS 1hjp 
HOLLIDAY JUNCTION BINDING PROTEIN RUVA FROM E. COLI 1hqm 
CRYSTAL STRUCTURE OF THERMUS AQUATICUS CORE RNA POLYMERASE-INCLUDES COMPLETE STRUCTURE WITH SIDE-CHAINS (EXCEPT FOR DISORDERED REGIONS)-FURTHER REFINED FROM ORIGINAL DEPOSITION-CONTAINS ADDITIONAL SEQUENCE INFORMATION 1huo 
CRYSTAL STRUCTURE OF DNA POLYMERASE BETA COMPLEXED WITH DNA AND CR-TMPPCP 1huz 
CRYSTAL STRUCTURE OF DNA POLYMERASE COMPLEXED WITH DNA AND CR-PCP 1i6v 
THERMUS AQUATICUS CORE RNA POLYMERASE-RIFAMPICIN COMPLEX 1iw7 
Crystal structure of the RNA polymerase holoenzyme from Thermus thermophilus at 2.6A resolution 1ixr 
RuvA-RuvB complex 1jj2 
Fully Refined Crystal Structure of the Haloarcula marismortui Large Ribosomal Subunit at 2.4 Angstrom Resolution 1jn3 
FIDELITY PROPERTIES AND STRUCTURE OF M282L MUTATOR MUTANT OF DNA POLYMERASE: SUBTLE STRUCTURAL CHANGES INFLUENCE THE MECHANISM OF NUCLEOTIDE DISCRIMINATION 1k73 
Co-crystal Structure of Anisomycin Bound to the 50S Ribosomal Subunit 1k8a 
Co-crystal structure of Carbomycin A bound to the 50S ribosomal subunit of Haloarcula marismortui 1k9m 
Co-crystal structure of tylosin bound to the 50S ribosomal subunit of Haloarcula marismortui 1kc8 
Co-crystal Structure of Blasticidin S Bound to the 50S Ribosomal Subunit 1kd1 
Co-crystal Structure of Spiramycin bound to the 50S Ribosomal Subunit of Haloarcula marismortui 1kft 
Solution Structure of the C-Terminal domain of UvrC from E-coli 1kg2 
Crystal structure of the core fragment of MutY from E.coli at 1.2A resolution 1kg3 
Crystal structure of the core fragment of MutY from E.coli at 1.55A resolution 1kg4 
Crystal structure of the K142A mutant of E. coli MutY (core fragment) 1kg5 
Crystal structure of the K142Q mutant of E.coli MutY (core fragment) 1kg6 
Crystal structure of the K142R mutant of E.coli MutY (core fragment) 1kg7 
Crystal Structure of the E161A mutant of E.coli MutY (core fragment) 1kqj 
Crystal Structure of a Mutant of MutY Catalytic Domain 1kqs 
The Haloarcula marismortui 50S Complexed with a Pretranslocational Intermediate in Protein Synthesis 1l9u 
THERMUS AQUATICUS RNA POLYMERASE HOLOENZYME AT 4 A RESOLUTION 1l9z 
Thermus aquaticus RNA Polymerase Holoenzyme/Fork-Junction Promoter DNA Complex at 6.5 A Resolution 1m1k 
Co-crystal structure of azithromycin bound to the 50S ribosomal subunit of Haloarcula marismortui 1m90 
Co-crystal structure of CCA-Phe-caproic acid-biotin and sparsomycin bound to the 50S ribosomal subunit 1mq2 
Human DNA Polymerase Beta Complexed With Gapped DNA Containing an 8-oxo-7,8-dihydro-Guanine and dAMP 1mq3 
Human DNA Polymerase Beta Complexed With Gapped DNA Containing an 8-oxo-7,8-dihydro-Guanine Template Paired with dCTP 1mud 
CATALYTIC DOMAIN OF MUTY FROM ESCHERICHIA COLI, D138N MUTANT COMPLEXED TO ADENINE 1mun 
CATALYTIC DOMAIN OF MUTY FROM ESCHERICHIA COLI D138N MUTANT 1muy 
CATALYTIC DOMAIN OF MUTY FROM ESCHERICHIA COLI 1n8r 
Structure of large ribosomal subunit in complex with virginiamycin M 1nji 
Structure of chloramphenicol bound to the 50S ribosomal subunit 1nom 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7), 31-KD DOMAIN; SOAKED IN THE PRESENCE OF MNCL2 (5 MILLIMOLAR) 1nzp 
Solution Structure of the Lyase Domain of Human DNA Polymerase Lambda 1orn 
Structure of a Trapped Endonuclease III-DNA Covalent Intermediate: Estranged-Guanine Complex 1orp 
Structure of a Trapped Endonuclease III-DNA Covalent Intermediate: Estranged-Adenine Complex 1p59 
Structure of a non-covalent Endonuclease III-DNA Complex 1pzn 
Rad51 (RadA) 1q7y 
Crystal Structure of CCdAP-Puromycin bound at the Peptidyl transferase center of the 50S ribosomal subunit 1q81 
Crystal Structure of minihelix with 3' puromycin bound to A-site of the 50S ribosomal subunit. 1q82 
Crystal Structure of CC-Puromycin bound to the A-site of the 50S ribosomal subunit 1q86 
Crystal structure of CCA-Phe-cap-biotin bound simultaneously at half occupancy to both the A-site and P-site of the the 50S ribosomal Subunit. 1qvf 
Structure of a deacylated tRNA minihelix bound to the E site of the large ribosomal subunit of Haloarcula marismortui 1qvg 
Structure of CCA oligonucleotide bound to the tRNA binding sites of the large ribosomal subunit of Haloarcula marismortui 1rpl 
2.3 ANGSTROMS CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF DNA POLYMERASE BETA 1rzt 
Crystal structure of DNA polymerase lambda complexed with a two nucleotide gap DNA molecule 1s72 
REFINED CRYSTAL STRUCTURE OF THE HALOARCULA MARISMORTUI LARGE RIBOSOMAL SUBUNIT AT 2.4 ANGSTROM RESOLUTION 1smy 
Structural basis for transcription regulation by alarmone ppGpp 1szp 
A Crystal Structure of the Rad51 Filament 1t4g 
ATPase in complex with AMP-PNP 1taq 
STRUCTURE OF TAQ DNA POLYMERASE 1tau 
TAQ POLYMERASE (E.C.2.7.7.7)/DNA/B-OCTYLGLUCOSIDE COMPLEX 1tv9 
HUMAN DNA POLYMERASE BETA COMPLEXED WITH NICKED DNA CONTAINING A MISMATCHED TEMPLATE ADENINE AND INCOMING CYTIDINE 1tva 
HUMAN DNA POLYMERASE BETA COMPLEXED WITH NICKED DNA CONTAINING A MISMATCHED TEMPLATE THYMIDINE AND INCOMING CYTIDINE 1v9p 
Crystal Structure Of Nad+-Dependent DNA Ligase 1vdd 
Crystal structure of recombinational repair protein RecR 1vq4 
The structure of the transition state analogue ""DAA"" bound to the large ribosomal subunit of Haloarcula marismortui 1vq5 
The structure of the transition state analogue ""RAA"" bound to the large ribosomal subunit of haloarcula marismortui 1vq6 
The structure of c-hpmn and CCA-PHE-CAP-BIO bound to the large ribosomal subunit of haloarcula marismortui 1vq7 
The structure of the transition state analogue ""DCA"" bound to the large ribosomal subunit of haloarcula marismortui 1vq8 
The structure of CCDA-PHE-CAP-BIO and the antibiotic sparsomycin bound to the large ribosomal subunit of haloarcula marismortui 1vq9 
The structure of CCA-PHE-CAP-BIO and the antibiotic sparsomycin bound to the large ribosomal subunit of haloarcula marismortui 1vqk 
The structure of CCDA-PHE-CAP-BIO bound to the a site of the ribosomal subunit of haloarcula marismortui 1vql 
The structure of the transition state analogue ""DCSN"" bound to the large ribosomal subunit of haloarcula marismortui 1vqm 
The structure of the transition state analogue ""DAN"" bound to the large ribosomal subunit of haloarcula marismortui 1vqn 
The structure of CC-HPMN AND CCA-PHE-CAP-BIO bound to the large ribosomal subunit of haloarcula marismortui 1vqo 
The structure of CCPMN bound to the large ribosomal subunit haloarcula marismortui 1vqp 
The structure of the transition state analogue ""RAP"" bound to the large ribosomal subunit of haloarcula marismortui 1w2b 
Trigger Factor ribosome binding domain in complex with 50S 1wef 
Catalytic Domain Of Muty From Escherichia Coli K20A Mutant 1weg 
Catalytic Domain Od Muty Form Escherichia Coli K142A Mutant 1wei 
Catalytic Domain Of Muty From Escherichia Coli K20A Mutant Complexed To Adenine 1x2i 
Crystal Structure Of Archaeal Xpf/Mus81 Homolog, Hef From Pyrococcus Furiosus, Helix-hairpin-helix Domain 1xsl 
Crystal Structure of human DNA polymerase lambda in complex with a one nucleotide DNA gap 1xsn 
Crystal Structure of human DNA polymerase lambda in complex with a one nucleotide DNA gap and ddTTP 1xsp 
Crystal Structure of human DNA polymerase lambda in complex with nicked DNA and pyrophosphate 1xu4 
ATPASE IN COMPLEX WITH AMP-PNP, MAGNESIUM AND POTASSIUM CO-F 1yhq 
Crystal Structure Of Azithromycin Bound To The G2099A Mutant 50S Ribosomal Subunit Of Haloarcula Marismortui 1yi2 
Crystal Structure Of Erythromycin Bound To The G2099A Mutant 50S Ribosomal Subunit Of Haloarcula Marismortui 1yij 
Crystal Structure Of Telithromycin Bound To The G2099A Mutant 50S Ribosomal Subunit Of Haloarcula Marismortui 1yit 
Crystal Structure Of Virginiamycin M and S Bound To The 50S Ribosomal Subunit Of Haloarcula Marismortui 1yj9 
Crystal Structure Of The Mutant 50S Ribosomal Subunit Of Haloarcula Marismortui Containing a three residue deletion in L22 1yjn 
Crystal Structure Of Clindamycin Bound To The G2099A Mutant 50S Ribosomal Subunit Of Haloarcula Marismortui 1yjw 
Crystal Structure Of Quinupristin Bound To The G2099A Mutant 50S Ribosomal Subunit Of Haloarcula Marismortui 1ynj 
Taq RNA polymerase-Sorangicin complex 1ynn 
Taq RNA polymerase-rifampicin complex 1z00 
Solution structure of the C-terminal domain of ERCC1 complexed with the C-terminal domain of XPF 1zjm 
Human DNA Polymerase beta complexed with DNA containing an A-A mismatched primer terminus 1zjn 
Human DNA Polymerase beta complexed with DNA containing an A-A mismatched primer terminus with dGTP 1zqa 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF KCL (150 MILLIMOLAR) AT PH 7.5 1zqb 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF BACL2 (150 MILLIMOLAR) 1zqc 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF CACL2 (15 MILLIMOLAR) 1zqd 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF CACL2 (150 MILLIMOLAR) 1zqe 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF CRCL3 (SATURATED SOLUTION) 1zqf 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF CSCL (150 MILLIMOLAR) 1zqg 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF A SODIUM-FREE ARTIFICIAL MOTHER LIQUOR AT PH 6.5 1zqh 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF A SODIUM-FREE ARTIFICIAL MOTHER LIQUOR AT PH 7.5 1zqi 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF KCL (150 MILLIMOLAR) 1zqj 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF CACL2 (15 MILLIMOLAR) AND MGCL2 (15 MILLIMOLAR) 1zqk 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF KCL (75 MILLIMOLAR) AND MGCL2 (75 MILLIMOLAR) 1zql 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF MNCL2 (15 MILLIMOLAR) AND MGCL2 (15 MILLIMOLAR) 1zqm 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF MNCL2 (15 MILLIMOLAR) 1zqn 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF BACL2 (15 MILLIMOLAR) AND NACL (15 MILLIMOLAR) 1zqo 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF CACL2 (15 MILLIMOLAR) AND NACL (15 MILLIMOLAR) 1zqp 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF KCL (75 MILLIMOLAR) AND NACL (75 MILLIMOLAR) 1zqq 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF MNCL2 (15 MILLIMOLAR) AND NACL (15 MILLIMOLAR) 1zqr 
DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX, SOAKED IN THE PRESENCE OF NICL2 1zqs 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF TLCL (0.5 MILLIMOLAR) 1zqt 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DATP (0.01 MILLIMOLAR) AND ZNCL2 (0.02 MILLIMOLAR) 1zqu 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7), 31-KD DOMAIN; SOAKED IN THE PRESENCE OF ARTIFICIAL MOTHER LIQUOR 1zqv 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7), 31-KD DOMAIN; SOAKED IN THE PRESENCE OF CACL2 (150 MILLIMOLAR) 1zqw 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7), 31-KD DOMAIN; SOAKED IN THE PRESENCE OF CSCL (150 MILLIMOLAR) 1zqx 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7), 31-KD DOMAIN; SOAKED IN THE PRESENCE OF KCL (150 MILLIMOLAR) 1zqy 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7), 31-KD DOMAIN; SOAKED IN THE PRESENCE OF MGCL2 (50 MILLIMOLAR) 1zqz 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7), 31-KD DOMAIN; SOAKED IN THE PRESENCE OF MNCL2 (50 MILLIMOLAR) 1zyr 
Structure of Thermus thermophilus RNA polymerase holoenzyme in complex with the antibiotic streptolydigin 2a1j 
Crystal Structure of the Complex between the C-Terminal Domains of Human XPF and ERCC1 2a68 
Crystal structure of the T. thermophilus RNA polymerase holoenzyme in complex with antibiotic rifabutin 2a69 
Crystal structure of the T. Thermophilus RNA polymerase holoenzyme in complex with antibiotic rifapentin 2a6e 
Crystal structure of the T. Thermophilus RNA polymerase holoenzyme 2a6h 
Crystal structure of the T. thermophilus RNA polymerase holoenzyme in complex with antibiotic sterptolydigin 2abk 
REFINEMENT OF THE NATIVE STRUCTURE OF ENDONUCLEASE III TO A RESOLUTION OF 1.85 ANGSTROM 2b21 
RADA Recombinase in complex with AMPPNP at pH 6.0 2bcq 
DNA polymerase lambda in complex with a DNA duplex containing an unpaired Dtmp 2bcr 
DNA polymerase lambda in complex with a DNA duplex containing an unpaired Damp 2bcs 
DNA polymerase lambda in complex with a DNA duplex containing an unpaired Dcmp 2bcu 
DNA polymerase lambda in complex with a DNA duplex containing an unpaired Damp and a T:T mismatch 2bcv 
DNA polymerase lambda in complex with Dttp and a DNA duplex containing an unpaired Dtmp 2be5 
Crystal structure of the T. Thermophilus RNA polymerase holoenzyme in complex with inhibitor tagetitoxin 2bgw 
XPF from Aeropyrum pernix, complex with DNA 2bhn 
XPF from Aeropyrum pernix 2bpc 
CRYSTAL STRUCTURE OF RAT DNA POLYMERASE BETA: EVIDENCE FOR A COMMON POLYMERASE MECHANISM 2bpf 
STRUCTURES OF TERNARY COMPLEXES OF RAT DNA POLYMERASE BETA, A DNA TEMPLATE-PRIMER, AND DDCTP 2bpg 
STRUCTURES OF TERNARY COMPLEXES OF RAT DNA POLYMERASE BETA, A DNA TEMPLATE-PRIMER, AND DDCTP 2csb 
Crystal structure of Topoisomerase V from Methanopyrus kandleri (61 kDa fragment) 2csd 
Crystal structure of Topoisomerase V (61 kDa fragment) 2cw0 
Crystal structure of Thermus thermophilus RNA polymerase holoenzyme at 3.3 angstroms resolution 2duy 
Crystal structure of competence protein ComEA-related protein from Thermus thermophilus HB8 2edu 
Solution structure of RSGI RUH-070, a C-terminal domain of kinesin-like protein KIF22 from human cDNA 2f1h 
RECOMBINASE IN COMPLEX WITH AMP-PNP and Potassium 2f1i 
Recombinase in Complex with AMP-PNP 2f1j 
Recombinase in Complex with ADP 2fmp 
DNA Polymerase beta with a terminated gapped DNA substrate and ddCTP with sodium in the catalytic site 2fmq 
Sodium in active site of DNA Polymerase Beta 2fms 
DNA Polymerase beta with a gapped DNA substrate and dUMPNPP with magnesium in the catalytic site 2fpk 
RadA recombinase in complex with ADP 2fpl 
RadA recombinase in complex with AMP-PNP and low concentration of K+ 2fpm 
RadA recombinase in complex with AMP-PNP and high concentration of K+ 2gho 
Recombinant Thermus aquaticus RNA polymerase for Structural Studies 2gws 
Crystal Structure of human DNA Polymerase lambda with a G/G mismatch in the primer terminus 2h56 
Crystal structure of DNA-3-methyladenine glycosidase (10174367) from Bacillus halodurans at 2.55 A resolution 2h5x 
RuvA from Mycobacterium tuberculosis 2i1q 
RadA Recombinase in complex with Calcium 2i9g 
DNA Polymerase Beta with a Benzo[c]phenanthrene diol epoxide adducted guanine base 2iso 
Ternary complex of DNA Polymerase beta with a dideoxy terminated primer and 2'-deoxyguanosine 5'-beta, gamma-difluoromethylene triphosphate 2isp 
Ternary complex of DNA Polymerase beta with a dideoxy terminated primer and 2'-deoxyguanosine 5'-beta, gamma-methylene triphosphate 2jhj 
3-methyladenine dna-glycosylase from Archaeoglobus fulgidus 2jhn 
3-methyladenine dna-glycosylase from Archaeoglobus fulgidus 2mut 
2MUT 2nrt 
Crystal structure of the C-terminal half of UvrC 2nrv 
Crystal structure of the C-terminal half of UvrC 2nrw 
Crystal structure of the C terminal half of UvrC 2nrx 
Crystal structure of the C-terminal half of UvrC, in the presence of sulfate molecules 2nrz 
Crystal structure of the C-terminal half of UvrC bound to its catalytic divalent cation 2o5i 
Crystal structure of the T. thermophilus RNA polymerase elongation complex 2o5j 
Crystal structure of the T. thermophilus RNAP polymerase elongation complex with the NTP substrate analog 2oce 
Crystal structure of Tex family protein PA5201 from Pseudomonas aeruginosa 2otj 
13-deoxytedanolide bound to the large subunit of Haloarcula marismortui 2otl 
Girodazole bound to the large subunit of Haloarcula marismortui 2owo 
Last Stop on the Road to Repair: Structure of E.coli DNA Ligase Bound to Nicked DNA-Adenylate 2p66 
Human DNA Polymerase beta complexed with tetrahydrofuran (abasic site) containing DNA 2p6r 
Crystal structure of superfamily 2 helicase Hel308 in complex with unwound DNA 2p6u 
Apo structure of the Hel308 superfamily 2 helicase 2pfn 
Na in the active site of DNA Polymerase lambda 2pfo 
DNA Polymerase lambda in complex with DNA and dUPNPP 2pfp 
DNA Polymerase lambda in complex with DNA and dCTP 2pfq 
Manganese promotes catalysis in a DNA polymerase lambda-DNA crystal 2ppb 
Crystal structure of the T. thermophilus RNAP polymerase elongation complex with the ntp substrate analog and antibiotic streptolydigin 2pxi 
Ternary complex of DNA polymerase beta with a dideoxy terminated primer and 2'-deoxyguanosine 5'-beta, gamma-monofluoromethylene triphosphate 2qa4 
A more complete structure of the the L7/L12 stalk of the Haloarcula marismortui 50S large ribosomal subunit 2qex 
Negamycin Binds to the Wall of the Nascent Chain Exit Tunnel of the 50S Ribosomal Subunit 2v1c 
Crystal structure and mutational study of RecOR provide insight into its role in DNA repair 2va8 
DNA Repair Helicase Hel308 2van 
Crystal structure of DNA polymerase beta 2w9m 
Structure of family X DNA polymerase from Deinococcus radiodurans 2yg8 
Structure of an unusual 3-METHYLADENINE DNA GLYCOSYLASE II (ALKA) FROM DEINOCOCCUS RADIODURANS 2yg9 
Structure of an unusual 3-METHYLADENINE DNA GLYCOSYLASE II (ALKA) FROM DEINOCOCCUS RADIODURANS 2ziu 
Crystal structure of the Mus81-Eme1 complex 2ziv 
Crystal structure of the Mus81-Eme1 complex 2ziw 
Crystal structure of the Mus81-Eme1 complex 2zix 
Crystal structure of the Mus81-Eme1 complex 2zj2 
Archaeal DNA helicase Hjm apo state in form 1 2zj5 
Archaeal DNA helicase Hjm complexed with ADP in form 1 2zj8 
Archaeal DNA helicase Hjm apo state in form 2 2zja 
Archaeal DNA helicase Hjm complexed with AMPPCP in form 2 2ztc 
MtRuvA Form II 2ztd 
MtRuvA Form III 2zte 
MtRuvA Form IV 3aoh 
RNA polymerase-Gfh1 complex (Crystal type 1) 3aoi 
RNA polymerase-Gfh1 complex (Crystal type 2) 3au2 
DNA polymerase X from Thermus thermophilus HB8 complexed with Ca-dGTP 3au6 
DNA polymerase X from Thermus thermophilus HB8 ternary complex with primer/template DNA and ddGTP 3auo 
DNA polymerase X from Thermus thermophilus HB8 ternary complex with 1-nt gapped DNA and ddGTP 3b0x 
K263A mutant of PolX from Thermus thermophilus HB8 complexed with Ca-dGTP 3b0y 
K263D mutant of PolX from Thermus thermophilus HB8 complexed with Ca-dGTP 3bzc 
Crystal Structure of the Tex protein from Pseudomonas aeruginosa, crystal form I 3bzk 
Crystal Structure of the Tex protein from Pseudomonas aeruginosa, crystal form 2 3c1y 
Structure of bacterial DNA damage sensor protein with co-purified and co-crystallized ligand 3c1z 
Structure of the ligand-free form of a bacterial DNA damage sensor protein 3c21 
Structure of a bacterial DNA damage sensor protein with reaction product 3c23 
Structure of a bacterial DNA damage sensor protein with non-reactive Ligand 3c2k 
DNA POLYMERASE BETA with a gapped DNA substrate and DUMPNPP with Manganese in the active site 3c2l 
Ternary complex of DNA POLYMERASE BETA with a C:DAPCPP mismatch in the active site 3c2m 
Ternary complex of DNA POLYMERASE BETA with a G:dAPCPP mismatch in the active site 3c5f 
Structure of a binary complex of the R517A Pol lambda mutant 3c5g 
Structure of a ternary complex of the R517K Pol lambda mutant 3c65 
Crystal Structure of Bacillus stearothermophilus UvrC 5' endonuclease domain 3cc2 
The Refined Crystal Structure of the Haloarcula Marismortui Large Ribosomal Subunit at 2.4 Angstrom Resolution with rrnA Sequence for the 23S rRNA and Genome-derived Sequences for r-Proteins 3cc4 
Co-crystal Structure of Anisomycin Bound to the 50S Ribosomal Subunit 3cc7 
Structure of Anisomycin resistant 50S Ribosomal Subunit: 23S rRNA mutation C2487U 3cce 
Structure of Anisomycin resistant 50S Ribosomal Subunit: 23S rRNA mutation U2535A 3ccj 
Structure of Anisomycin resistant 50S Ribosomal Subunit: 23S rRNA mutation C2534U 3ccl 
Structure of Anisomycin resistant 50S Ribosomal Subunit: 23S rRNA mutation U2535C. Density for Anisomycin is visible but not included in model. 3ccm 
Structure of Anisomycin resistant 50S Ribosomal Subunit: 23S rRNA mutation G2611U 3ccq 
Structure of Anisomycin resistant 50S Ribosomal Subunit: 23S rRNA mutation A2488U 3ccr 
Structure of Anisomycin resistant 50S Ribosomal Subunit: 23S rRNA mutation A2488C. Density for anisomycin is visible but not included in the model. 3ccs 
Structure of Anisomycin resistant 50S Ribosomal Subunit: 23S rRNA mutation G2482A 3ccu 
Structure of Anisomycin resistant 50S Ribosomal Subunit: 23S rRNA mutation G2482C 3ccv 
Structure of Anisomycin resistant 50S Ribosomal Subunit: 23S rRNA mutation G2616A 3cd6 
Co-cystal of large Ribosomal Subunit mutant G2616A with CC-Puromycin 3cma 
The structure of CCA and CCA-Phe-Cap-Bio bound to the large ribosomal subunit of Haloarcula marismortui 3cme 
The Structure of CA and CCA-PHE-CAP-BIO Bound to the Large Ribosomal Subunit of Haloarcula Marismortui 3cpw 
The structure of the antibiotic LINEZOLID bound to the large ribosomal subunit of HALOARCULA MARISMORTUI 3cxc 
The structure of an enhanced oxazolidinone inhibitor bound to the 50S ribosomal subunit of H. marismortui 3dxj 
Crystal structure of thermus thermophilus rna polymerase holoenzyme in complex with the antibiotic myxopyronin 3eql 
Crystal structure of the T. Thermophilus RNA polymerase holoenzyme in complex with antibiotic myxopyronin 3etl 
RadA recombinase from Methanococcus maripaludis in complex with AMPPNP 3ew9 
RADA recombinase from METHANOCOCCUS MARIPALUDIS in complex with AMPPNP and potassium ions 3ewa 
RADA recombinase from METHANOCOCCUS MARIPALUDIS in complex with AMPPNP and ammonium ions 3f0z 
Crystal structure of Clostridium acetobutylicum 8-oxoguanine glycosylase/lyase in its apo-form 3fyh 
Recombinase in complex with ADP and metatungstate 3gdx 
Dna polymerase beta with a gapped DND substrate and dTMP(CF2)PP 3hw8 
ternary complex of DNA polymerase lambda of a two nucleotide gapped DNA substrate with a C in the scrunch site 3hwt 
Ternary complex of DNA polymerase lambda bound to a two nucleotide gapped DNA substrate with a scrunched dA 3hx0 
ternary complex of L277A, H511A, R514 mutant pol lambda bound to a 2 nucleotide gapped DNA substrate with a scrunched dA 3i55 
Co-crystal structure of Mycalamide A Bound to the Large Ribosomal Subunit 3i56 
Co-crystal structure of Triacetyloleandomcyin Bound to the Large Ribosomal Subunit 3isb 
Binary complex of human DNA polymerase beta with a gapped DNA 3isc 
Binary complex of human DNA polymerase beta with an abasic site (THF) in the gapped DNA 3isd 
Ternary complex of human DNA polymerase beta with an abasic site (THF): DAPCPP mismatch 3jpn 
Ternary complex of DNA polymerase beta with a dideoxy terminated primer and 2'-deoxyguanosine 5'-beta, gamma-dichloro methylene triphosphate 3jpo 
Ternary complex of DNA polymerase beta with a dideoxy terminated primer and 2'-deoxyguanosine 5'-beta, gamma-monochloromethylene triphosphate 3jpp 
Ternary complex of DNA polymerase beta with a dideoxy terminated primer and 2'-deoxyguanosine 5'-beta, gamma-MonoMethyl Methylene triphosphate 3jpq 
Ternary complex of DNA polymerase beta with a dideoxy terminated primer and 2'-deoxyguanosine 5'-beta, gamma-monoBromo methylene triphosphate 3jpr 
Ternary complex of DNA polymerase beta with a dideoxy terminated primer and 2'-deoxyguanosine 5'-beta, gamma-dimethyl methylene triphosphate 3jps 
Ternary complex of DNA polymerase beta with a dideoxy terminated primer and 2'-deoxyguanosine 5'-beta, gamma-fluoro methyl methylene triphosphate 3jpt 
Ternary complex of DNA polymerase beta with a dideoxy terminated primer and 2'-deoxyguanosine 5'-beta, gamma-fluoro chloro methylene triphosphate 3k75 
X-ray crystal structure of reduced XRCC1 bound to DNA pol beta catalytic domain 3lda 
Yeast Rad51 H352Y Filament Interface Mutant 3lk9 
DNA polymerase beta with a gapped DNA substrate and dTMP(CF2)P(CF2)P 3maj 
Crystal structure of putative DNA processing protein DprA from Rhodopseudomonas palustris CGA009 3mby 
Ternary complex of DNA Polymerase BETA with template base A and 8oxodGTP in the active site with a dideoxy terminated primer 3mda 
DNA polymerase lambda in complex with araC 3mdc 
DNA polymerase lambda in complex with dFdCTP 3mgh 
Binary complex of a DNA polymerase lambda loop mutant 3mgi 
Ternary complex of a DNA polymerase lambda loop mutant 3ntu 
RADA RECOMBINASE D302K MUTANT IN COMPLEX with AMP-PNP 3ogu 
DNA Polymerase beta mutant 5P20 complexed with 6bp of DNA 3pml 
crystal structure of a polymerase lambda variant with a dGTP analog opposite a templating T 3pmn 
ternary crystal structure of polymerase lambda variant with a GT mispair at the primer terminus with Mn2+ in the active site 3pnc 
Ternary crystal structure of a polymerase lambda variant with a GT mispair at the primer terminus and sodium at catalytic metal site 3rh4 
DNA Polymerase Beta with a dideoxy-terminated primer with an incoming ribonucleotide (rCTP) 3rh5 
DNA Polymerase Beta Mutant (Y271) with a dideoxy-terminated primer with an incoming deoxynucleotide (dCTP) 3rh6 
DNA Polymerase Beta Mutant (Y271) with a dideoxy-terminated primer with an incoming ribonucleotide (rCTP) 3rje 
Ternary complex of DNA Polymerase Beta with a gapped DNA containing 8odG at template position 3rjf 
Ternary complex of DNA Polymerase Beta with a gapped DNA containing (syn)8odG at template position paired with non-hydrolyzable dATP analog (dApCPP) 3rjg 
Binary complex of DNA Polymerase Beta with a gapped DNA containing 8odG:dA base-pair at primer Terminus 3rjh 
Ternary complex of DNA Polymerase Beta with a gapped DNA containing (syn)8odG:dA at primer terminus and dG:dCMP(CF2)PPin the active site 3rji 
Ternary complex of DNA Polymerase Beta with a gapped DNA containing 8odG at template position paired with non-hydrolyzable dCTP analog (dCMP(CF2)PP) 3rjj 
Ternary complex crystal structure of DNA Polymerase Beta with template 8odG provides insight into mutagenic lesion bypass 3rjk 
Ternary complex of DNA Polymerase Beta with a gapped DNA containing 8odG:dC base pair at primer terminus and dG:dCMP(CF2)PP in the active site 3s6i 
Schizosaccaromyces pombe 3-methyladenine DNA glycosylase (Mag1) in complex with abasic-DNA. 3sgi 
Crystal structure of DNA ligase A BRCT domain deleted mutant of Mycobacterium tuberculosis 3tfr 
Ternary complex structure of DNA polymerase beta with a gapped DNA substrate and a, b dAMP(CF2)PP in the active site 3tfs 
Ternary complex structure of DNA polymerase beta with a gapped DNA substrate and a, b dAMP(CFH)PP in the active site: Stereoselective binding of (S) isomer 3upq 
Crystal structure of the pre-catalytic ternary complex of polymerase lambda with an rATP analog opposite a templating T. 3uq0 
Crystal structure of the post-catalytic product complex of polymerase lambda with an rAMP at the primer terminus. 3uq2 
Crystal structure of the post-catalytic product complex of polymerase lambda with an rCMP inserted opposite a templating G and dAMP inserted opposite a templating T at the primer terminus. 3uxn 
Crystal Structure of Rat DNA Polymerase Beta, Wild Type Apoenzyme 3uxo 
Crystal Structure of Rat DNA Polymerase Beta Mutator I260Q Apoenzyme 3uxp 
Co-crystal Structure of Rat DNA polymerase beta Mutator I260Q: Enzyme-DNA-ddTTP 3v72 
Crystal Structure of Rat DNA polymerase beta Mutator E295K: Enzyme-dsDNA 3v7j 
Co-crystal structure of Wild Type Rat polymerase beta: Enzyme-DNA binary complex 3v7k 
Co-crystal structure of K72E variant of rat polymerase beta: Enzyme-DNA binary complex 3v7l 
Apo Structure of Rat DNA polymerase beta K72E variant 3vdp 
Structure and biochemical studies of the recombination mediator protein RecR in RecFOR pathway 3vdu 
Structure of recombination mediator protein RecRK21G mutant 3wod 
RNA polymerase-gp39 complex 3zd8 
Potassium bound structure of E. coli ExoIX in P1 3zd9 
Potassium bound structure of E. coli ExoIX in P21 3zda 
Structure of E. coli ExoIX in complex with a fragment of the Flap1 DNA oligonucleotide, potassium and magnesium 3zdb 
Structure of E. coli ExoIX in complex with the palindromic 5ov4 DNA oligonucleotide, di-magnesium and potassium 3zdc 
Structure of E. coli ExoIX in complex with the palindromic 5ov4 DNA oligonucleotide, potassium and calcium 3zdd 
Structure of E. coli ExoIX in complex with the palindromic 5ov6 oligonucleotide and potassium 3zde 
Potassium free structure of E. coli ExoIX 4adx 
The Cryo-EM Structure of the Archaeal 50S Ribosomal Subunit in Complex with Initiation Factor 6 4ak9 
Structure of chloroplast FtsY from Physcomitrella patens 4do9 
Ternary complex of dna polymerase beta with a dideoxy terminated primer and 2'-deoxyguanosine 5'-beta, gamma-monofluoromethylene triphosphate: stereoselective binding of r-isomer 4doa 
Ternary complex of dna polymerase beta with a dideoxy terminated primer and 2'-deoxyguanosine 5'-beta, gamma-monofluoromethylene triphosphate: non-interactive binding of s-isomer 4dob 
Ternary complex of DNA polymerase beta with a dideoxy terminated primer and 2'-deoxyguanosine 5'-beta, gamma-monochlororomethylene triphosphate: Stereoselective binding of R-isomer 4doc 
Ternary complex of dna polymerase beta with a dideoxy terminated primer and 2'-deoxyguanosine 5'-beta, gamma-monochlororomethylene triphosphate:binding of S-isomer 4ejy 
Structure of MBOgg1 in complex with high affinity DNA ligand 4ejz 
Structure of MBOgg1 in complex with low affinity DNA ligand 4f5n 
Open ternary complex of R283K DNA polymerase beta with a metal free dCTP analog 4f5o 
Open ternary complex of R283K DNA polymerase beta with a one metal bound dCTP analog 4f5p 
Open ternary mismatch complex of R283K DNA polymerase beta with a dATP analog 4f5q 
Closed ternary complex of R283K DNA polymerase beta 4f5r 
Open and closed ternary complex of R283K DNA polymerase beta with a dCTP analog in the same asymmetric unit 4fo6 
Crystal structure of the pre-catalytic ternary complex of polymerase lambda with a dATP analog opposite a templating T and an rCMP at the primer terminus. 4g7h 
Crystal structure of Thermus thermophilus transcription initiation complex 4g7o 
Crystal structure of Thermus thermophilus transcription initiation complex containing 2 nt of RNA 4g7z 
Crystal structure of Thermus thermophilus transcription initiation complex containing 5-BrU at template-strand position +1 4gfj 
Crystal structure of Topo-78, an N-terminal 78kDa fragment of topoisomerase V 4glx 
DNA ligase A in complex with inhibitor 4gxi 
R283K DNA polymerase beta binary complex with a templating 8OG 4gxj 
R283K DNA polymerase beta ternary complex with a templating 8OG and incoming dCTP analog 4gxk 
R283K DNA polymerase beta ternary complex with a templating 8OG and incoming dATP analog 4gzy 
Crystal structures of bacterial RNA Polymerase paused elongation complexes 4gzz 
Crystal structures of bacterial RNA Polymerase paused elongation complexes 4itq 
Crystal structure of hypothetical protein SCO1480 bound to DNA 4jcv 
Crystal structure of the RecOR complex in an open conformation 4jwm 
Ternary complex of D256E mutant of DNA Polymerase Beta 4jwn 
Ternary complex of D256A mutant of DNA Polymerase Beta 4k4g 
4K4G 4k4h 
4K4H 4k4i 
4K4I 4kld 
DNA polymerase beta matched substrate complex with Ca2+, 0 s 4kle 
DNA polymerase beta matched reactant complex with Mg2+, 10 s 4klf 
DNA polymerase beta matched reactant complex with Mg2+, 20 s 4klg 
DNA polymerase beta matched product complex with Mg2+, 40 s 4klh 
DNA polymerase beta matched product complex with Mn2+, 40 s 4kli 
DNA polymerase beta matched product complex with Mg2+, 90 s 4klj 
DNA polymerase beta matched product complex with Mg2+, 5 min 4kll 
DNA polymerase beta matched product complex with Mg2+, 45 min 4klm 
DNA polymerase beta matched product complex with Mg2+, 11 h 4klo 
DNA polymerase beta matched nick complex with Mg2+ and PPi, 30 min 4klq 
Observing a DNA polymerase choose right from wrong. 4kls 
DNA polymerase beta mismatched reactant complex with Mn2+, 10 min 4klt 
DNA polymerase beta mismatched product complex with Mn2+, 30 min 4klu 
DNA polymerase beta mismatched product complex with Mn2+, 15 h 4lvs 
DNA polymerase beta mismatched substrate complex with Mn2+, 2.5 min 4m2y 
Structure of human DNA polymerase beta complexed with 8-BrG as the template base in a 1-nucleotide gapped DNA 4m47 
structure of human DNA polymerase complexed with 8-BrG in the template base paired with incoming non-hydrolyzable GTP 4m9g 
DNA Polymerase Beta E295K Binary Complex 4m9h 
DNA Polymerase Beta E295K Soaked with dTTP 4m9j 
DNA Polymerase Beta E295K Soaked with dUMPNPP 4m9l 
DNA Polymerase Beta E295K Soaked with dCTP 4m9n 
DNA Polymerase Beta E295K Soaked with dATP 4mf2 
Structure of human DNA polymerase beta complexed with O6MG as the template base in a 1-nucleotide gapped DNA 4mf8 
4MF8 4mfa 
4MFA 4mfc 
Structure of human DNA polymerase beta complexed with O6MG in the template base paired with incoming non-hydrolyzable CTP 4mff 
Structure of human DNA polymerase beta complexed with O6MG in the template base paired with incoming non-hydrolyzable TTP 4mq9 
Crystal structure of Thermus thermophilus RNA polymerase holoenzyme in complex with GE23077 4nlk 
Structure of human DNA polymerase beta complexed with 8BrG in the template base-paired with incoming non-hydrolyzable CTP 4nln 
Structure of human DNA polymerase beta complexed with nicked DNA containing a template 8BrG and incoming CTP 4nlz 
Structure of human DNA polymerase beta complexed with nicked DNA containing a mismatched template 8BrG and incoming GTP 4nm1 
Structure of human DNA polymerase beta complexed with a nicked DNA containing a 8BrG-C at N-1 position and G-C at N position 4nm2 
Structure of human DNA polymerase beta complexed with a nicked DNA containing a 8BrG-G at N-1 position and G-C at N position 4nxz 
DNA polymerase beta with O6mG in the template base opposite to incoming non-hydrolyzable TTP with manganese in the active site 4ny8 
DNA polymerase beta with O6mG in the template base opposite to incoming non-hydrolyzable CTP with manganese in the active site 4o5c 
4O5C 4o5e 
4O5E 4o5k 
4O5K 4o6o 
4O6O 4o6p 
4O6P 4o9m 
Human DNA polymerase beta complexed with adenylated tetrahydrofuran (abasic site) containing DNA 4oin 
Crystal structure of Thermus thermophilus transcription initiation complex soaked with GE23077 4oio 
Crystal structure of Thermus thermophilus pre-insertion substrate complex for de novo transcription initiation 4oip 
Crystal structure of Thermus thermophilus transcription initiation complex soaked with GE23077, ATP, and CMPcPP 4oiq 
Crystal structure of Thermus thermophilus transcription initiation complex soaked with GE23077 and rifampicin 4oir 
Crystal structure of Thermus thermophilus RNA polymerase transcription initiation complex soaked with GE23077 and rifamycin SV 4p0p 
4P0P 4p0q 
4P0Q 4p0r 
4P0R 4p0s 
4P0S 4p2h 
4P2H 4p4m 
4P4M 4p4o 
4P4O 4p4p 
4P4P 4pgq 
4PGQ 4pgx 
4PGX 4pgy 
4PGY 4ph5 
4PH5 4pha 
4PHA 4phd 
4PHD 4phe 
4PHE 4php 
4PHP 4ppx 
DNA Polymerase Beta E295K with Spiroiminodihydantoin in Templating Position 4q4z 
4Q4Z 4q5s 
4Q5S 4r63 
4R63 4r64 
4R64 4r65 
4R65 4r66 
4R66 4rpx 
4RPX 4rpy 
4RPY 4rpz 
4RPZ 4rq0 
4RQ0 4rq1 
4RQ1 4rq2 
4RQ2 4rq3 
4RQ3 4rq4 
4RQ4 4rq5 
4RQ5 4rq6 
4RQ6 4rq7 
4RQ7 4rq8 
4RQ8 4rt2 
4RT2 4rt3 
4RT3 4tup 
4TUP 4tuq 
4TUQ 4tur 
4TUR 4tus 
4TUS 4uaw 
4UAW 4uay 
4UAY 4uaz 
4UAZ 4ub1 
4UB1 4ub2 
4UB2 4ub3 
4UB3 4ub4 
4UB4 4ub5 
4UB5 4ubb 
4UBB 4ubc 
4UBC 4v9f 
4V9F 4wqs 
4WQS 4wqt 
4WQT 4x5v 
4X5V 4xa5 
4XA5 4xln 
4XLN 4xlp 
4XLP 4xlq 
4XLQ 4xlr 
4XLR 4xls 
4XLS 4xq8 
4XQ8 4xrh 
4XRH 4xus 
4XUS 4ymm 
4YMM 4ymn 
4YMN 4ymo 
4YMO 4yn4 
4YN4 4yvz 
4YVZ 4yxj 
4YXJ 4yxm 
4YXM 4z6c 
4Z6C 4z6d 
4Z6D 4z6e 
4Z6E 4z6f 
4Z6F 5bol 
5BOL 5bom 
5BOM 5bpc 
5BPC 5ca7 
5CA7 5cb1 
5CB1 5chg 
5CHG 5cj7 
5CJ7 5cp2 
5CP2 5cr0 
5CR0 5cwr 
5CWR 5d4c 
5D4C 5d4d 
5D4D 5d4e 
5D4E 5db6 
5DB6 5db7 
5DB7 5db8 
5DB8 5db9 
5DB9 5dba 
5DBA 5dbb 
5DBB 5dbc 
5DBC 5ddm 
5DDM 5ddy 
5DDY 5dkw 
5DKW 5e17 
5E17 5e18 
5E18 5eoz 
5EOZ 5hm5 
5HM5 5i2d 
5I2D 5iii 
5III 5iij 
5IIJ 5iik 
5IIK 5iil 
5IIL 5iim 
5IIM 5iin 
5IIN 5iio 
5IIO 5j0o 
5J0O 5j0p 
5J0P 5j0q 
5J0Q 5j0r 
5J0R 5j0s 
5J0S 5j0t 
5J0T 5j0u 
5J0U 5j0v 
5J0V 5j0w 
5J0W 5j0x 
5J0X 5j0y 
5J0Y 5j29 
5J29 5j2a 
5J2A 5j2b 
5J2B 5j2c 
5J2C 5j2d 
5J2D 5j2e 
5J2E 5j2f 
5J2F 5j2g 
5J2G 5j2h 
5J2H 5j2i 
5J2I 5j2j 
5J2J 5j2k 
5J2K 5jzc 
5JZC 5tmc 
5TMC 5tmf 
5TMF 7ice 
DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX, SOAKED IN THE PRESENCE OF CACL2 7icf 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SIX BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF CDCL2 (0.1 MILLIMOLAR) (FOUR-DAY SOAK) 7icg 
DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX, SOAKED IN THE PRESENCE OF CDCL2 7ich 
DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX, SOAKED IN THE PRESENCE OF COCL2 7ici 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SIX BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF CRCL3 (0.1 MILLIMOLAR) 7icj 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SIX BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF CUCL2 (0.1 MILLIMOLAR) 7ick 
DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX, SOAKED IN THE PRESENCE OF MGCL2 7icl 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SIX BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF MNCL2 (0.1 MILLIMOLAR) 7icm 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SIX BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF MNCL2 (1.0 MILLIMOLAR) 7icn 
DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX, SOAKED IN THE PRESENCE OF NICL2 7ico 
DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX, SOAKED IN THE PRESENCE OF ZNCL2 7icp 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SIX BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF ZNCL2 (0.01 MILLIMOLAR) 7icq 
DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX, SOAKED IN THE PRESENCE OF ZNCL2 7icr 
DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX, SOAKED IN THE PRESENCE OF ZNCL2 7ics 
DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX, SOAKED IN THE PRESENCE OF ZNCL2 7ict 
DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX, SOAKED IN THE PRESENCE OF ZNCL2 AND MGCL2 7icu 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SIX BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF CDCL2 (0.1 MILLIMOLAR) 7icv 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SIX BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF MNCL2 (0.1 MILLIMOLAR) AND IN THE ABSENCE OF NACL 8ica 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DATP (1 MILLIMOLAR) AND CACL2 (5 MILLIMOLAR) 8icb 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF ARTIFICIAL MOTHER LIQUOR 8icc 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA (NO 5'-PHOSPHATE) 8ice 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DATP (1 MILLIMOLAR) AND CDCL2 (1 MILLIMOLAR) 8icf 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DATP (10 MILLIMOLAR) AND MGCL2 (50 MILLIMOLAR) 8icg 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DATP (1 MILLIMOLAR) AND MGCL2 (5 MILLIMOLAR) 8ich 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DCTP (1 MILLIMOLAR) AND MGCL2 (5 MILLIMOLAR) 8ici 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DGTP (1 MILLIMOLAR) AND MGCL2 (5 MILLIMOLAR) 8icj 
DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX + THYMIDINE-5'-TRIPHOSPHATE, SOAKED IN THE PRESENCE OF DTTP AND MGCL2 8ick 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DATP (1 MILLIMOLAR), MGCL2 (5 MILLIMOLAR), AND MNCL2 (5 MILLIMOLAR) 8icl 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DATP (1 MILLIMOLAR) AND NICL2 (5 MILLIMOLAR) 8icm 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DATP (1 MILLIMOLAR), MNCL2 (5 MILLIMOLAR), AND AMMONIUM SULFATE (75 MILLIMOLAR) 8icn 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF ATP (1 MILLIMOLAR) AND MNCL2 (5 MILLIMOLAR) 8ico 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF AZT-TP (1 MILLIMOLAR) AND MNCL2 (5 MILLIMOLAR) 8icp 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DATP (1 MILLIMOLAR) AND MNCL2 (5 MILLIMOLAR) 8icq 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF OF DATP (0.1 MILLIMOLAR) AND MNCL2 (0.5 MILLIMOLAR) 8icr 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DATP (1 MILLIMOLAR) AND MNCL2 (5 MILLIMOLAR) 8ics 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DCTP (1 MILLIMOLAR) AND MNCL2 (5 MILLIMOLAR) 8ict 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DCTP (1 MILLIMOLAR) AND MNCL2 (5 MILLIMOLAR) 8icu 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DDATP (1 MILLIMOLAR) AND MNCL2 (5 MILLIMOLAR) 8icv 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DGTP (1 MILLIMOLAR) AND MNCL2 (5 MILLIMOLAR) 8icw 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DTTP (1 MILLIMOLAR) AND MNCL2 (5 MILLIMOLAR) 8icx 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DTTP (1 MILLIMOLAR) AND MNCL2 (5 MILLIMOLAR) 8icy 
DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX + THYMIDINE-5'-TRIPHOSPHATE, SOAKED IN THE PRESENCE OF DTTP AND MNCL2 8icz 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF OF DATP (1 MILLIMOLAR), MNCL2 (5 MILLIMOLAR), AND LITHIUM SULFATE (75 MILLIMOLAR) 9ica 
DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX + 2'-DEOXYADENOSINE-5'-O-(1-THIOTRIPHOSPHATE), SOAKED IN THE PRESENCE OF DATP(ALPHA)S AND MNCL2 9icb 
DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX + 2'-DEOXYADENOSINE-5'-TRIPHOSPHATE, SOAKED IN THE PRESENCE OF DATP AND COCL2 9icc 
DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX + 2'-DEOXYADENOSINE-5'-TRIPHOSPHATE, SOAKED IN THE PRESENCE OF DATP AND CRCL3 9ice 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DATP (1 MILLIMOLAR) AND CUCL2 (0.1 MILLIMOLAR) 9icf 
DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX + 2'-DEOXYADENOSINE-5'-TRIPHOSPHATE, SOAKED IN THE PRESENCE OF DATP AND ZNCL2 9icg 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DCTP (1 MILLIMOLAR) AND ZNCL2 (1 MILLIMOLAR) 9ich 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DGTP (1 MILLIMOLAR) AND ZNCL2 (1 MILLIMOLAR) 9ici 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DTTP (1 MILLIMOLAR) AND ZNCL2 (1 MILLIMOLAR) 9icj 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA 9ick 
DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX, SOAKED IN THE PRESENCE OF ARTIFICIAL MOTHER LIQUOR 9icl 
DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX, SOAKED IN THE PRESENCE OF PYROPHOSPHATE AND MNCL2 9icm 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SIX BASE PAIRS OF DOUBLE STRANDED DNA (NO 5'-PHOSPHATE) 9icn 
DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX + 2',3'-DIDEOXYCYTIDINE-5'-TRIPHOSPHATE, SOAKED IN THE PRESENCE OF DDCTP AND MGCL2 9ico 
DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX, SOAKED IN THE PRESENCE OF DTTP AND MGCL2 9icp 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SIX BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF PYROPHOSPHATE (1 MILLIMOLAR) AND MGCL2 (5 MILLIMOLAR) 9icq 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SIX BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DATP (1 MILLIMOLAR) AND MNCL2 (5 MILLIMOLAR) 9icr 
DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX + 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, SOAKED IN THE PRESENCE OF DCTP AND MNCL2 9ics 
DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX + 2',3'-DIDEOXYCYTIDINE-5'-TRIPHOSPHATE, SOAKED IN THE PRESENCE OF DDCTP AND MNCL2 9ict 
DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX + 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, SOAKED IN THE PRESENCE OF DGTP AND MNCL2 9icu 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SIX BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DTTP (1 MILLIMOLAR) AND MNCL2 (5 MILLIMOLAR) 9icv 
DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX + 2'-DEOXYADENOSINE-5'-TRIPHOSPHATE, SOAKED IN THE PRESENCE OF DATP AND ZNCL2 9icw 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SIX BASE PAIRS OF DNA; NATIVE STRUCTURE 9icx 
DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SIX BASE PAIRS OF DNA (NON GAPPED DNA ONLY) 9icy 
DNA POLYMERASE BETA (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA (NON GAPPED DNA ONLY) - Links (links to other resources describing this domain)
-
PFAM HHH INTERPRO IPR003583

