The domain within your query sequence starts at position 145 and ends at position 220; the E-value for the IG_like domain shown below is 4.02e-2.
TPKVTCVVVDVSEDDPDVRISWFVNNVEVHTAQTQTHREDYNSTIRVVSALPIQHQDWMS GKEFKCKVNNKDLPSP
IG_likeImmunoglobulin like |
|---|
| SMART accession number: | SM00410 |
|---|---|
| Description: | IG domains that cannot be classified into one of IGv1, IGc1, IGc2, IG. |
| Family alignment: |
There are 91644 IG_like domains in 55038 proteins in SMART's nrdb database.
Click on the following links for more information.
- Evolution (species in which this domain is found)
-
Taxonomic distribution of proteins containing IG_like domain.
This tree includes only several representative species. The complete taxonomic breakdown of all proteins with IG_like domain is also avaliable.
Click on the protein counts, or double click on taxonomic names to display all proteins containing IG_like domain in the selected taxonomic class.
- Metabolism (metabolic pathways involving proteins which contain this domain)
-
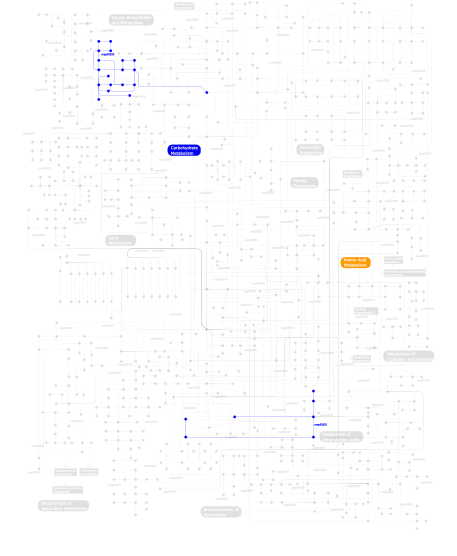
Click the image to view the interactive version of the map in iPath% proteins involved KEGG pathway ID Description 23.43 map04514 Cell adhesion molecules (CAMs) 11.54 map04060 Cytokine-cytokine receptor interaction 8.39 map04640 Hematopoietic cell lineage 6.99 map04662 B cell receptor signaling pathway 4.20 map04670 Leukocyte transendothelial migration 4.20 map04510 Focal adhesion 3.67 map04650 Natural killer cell mediated cytotoxicity 3.50 map04520 Adherens junction 3.32 map04010 MAPK signaling pathway 2.27 map04612 Antigen processing and presentation 2.27 map04360 Axon guidance 2.27 map05210 Colorectal cancer 2.27 map04940 Type I diabetes mellitus 2.10 map05221 Acute myeloid leukemia 1.75 map04210 Apoptosis 1.57 map04810 Regulation of actin cytoskeleton 1.57 map04020 Calcium signaling pathway 1.57 map04660 T cell receptor signaling pathway 1.40 map05214 Glioma 1.40 map04620 Toll-like receptor signaling pathway 1.40 map05215 Prostate cancer 1.40 map04540 Gap junction 1.40 map05218 Melanoma 1.22 map04370 VEGF signaling pathway 1.22 map04916 Melanogenesis 1.22  map00530
map00530Aminosugars metabolism 0.87 map04512 ECM-receptor interaction 0.52 map05120 Epithelial cell signaling in Helicobacter pylori infection 0.35 map04080 Neuroactive ligand-receptor interaction 0.17 map00970 Aminoacyl-tRNA biosynthesis 0.17  map00251
map00251Glutamate metabolism 0.17 map04920 Adipocytokine signaling pathway 0.17 map04530 Tight junction This information is based on mapping of SMART genomic protein database to KEGG orthologous groups. Percentage points are related to the number of proteins with IG_like domain which could be assigned to a KEGG orthologous group, and not all proteins containing IG_like domain. Please note that proteins can be included in multiple pathways, ie. the numbers above will not always add up to 100%.
- Structure (3D structures containing this domain)
3D Structures of IG_like domains in PDB
PDB code Main view Title 12e8 
2E8 FAB FRAGMENT 15c8 
CATALYTIC ANTIBODY 5C8, FREE FAB 1a0q 
29G11 COMPLEXED WITH PHENYL [1-(1-N-SUCCINYLAMINO)PENTYL] PHOSPHONATE 1a3l 
CATALYSIS OF A DISFAVORED REACTION: AN ANTIBODY EXO DIELS-ALDERASE-TSA-INHIBITOR COMPLEX AT 1.95 A RESOLUTION 1a3r 
FAB FRAGMENT (ANTIBODY 8F5) COMPLEXED WITH PEPTIDE FROM HUMAN RHINOVIRUS (SEROTYPE 2) VIRAL CAPSID PROTEIN VP2 (RESIDUES 156-170) 1a4j 
DIELS ALDER CATALYTIC ANTIBODY GERMLINE PRECURSOR 1a4k 
DIELS ALDER CATALYTIC ANTIBODY WITH TRANSITION STATE ANALOGUE 1a5f 
FAB FRAGMENT OF A MONOCLONAL ANTI-E-SELECTIN ANTIBODY 1a6p 
ENGINEERING OF A MISFOLDED FORM OF CD2 1a6t 
FAB FRAGMENT OF MAB1-IA MONOCLONAL ANTIBODY TO HUMAN RHINOVIRUS 14 NIM-IA SITE 1a8j 
IMMUNOGLOBULIN LAMBDA LIGHT CHAIN DIMER (MCG) COMPLEX WITH ASPARTAME 1acy 
CRYSTAL STRUCTURE OF THE PRINCIPAL NEUTRALIZING SITE OF HIV-1 1ad0 
FAB FRAGMENT OF ENGINEERED HUMAN MONOCLONAL ANTIBODY A5B7 1ad9 
IGG-FAB FRAGMENT OF ENGINEERED HUMAN MONOCLONAL ANTIBODY CTM01 1adq 
CRYSTAL STRUCTURE OF A HUMAN IGM RHEUMATOID FACTOR FAB IN COMPLEX WITH ITS AUTOANTIGEN IGG FC 1ae6 
IGG-FAB FRAGMENT OF MOUSE MONOCLONAL ANTIBODY CTM01 1afv 
HIV-1 CAPSID PROTEIN (P24) COMPLEX WITH FAB25.3 1ahw 
A COMPLEX OF EXTRACELLULAR DOMAIN OF TISSUE FACTOR WITH AN INHIBITORY FAB (5G9) 1ai1 
HIV-1 V3 LOOP MIMIC 1aif 
ANTI-IDIOTYPIC FAB 409.5.3 (IGG2A) FAB FROM MOUSE 1aj7 
IMMUNOGLOBULIN 48G7 GERMLINE FAB ANTIBODY COMPLEXED WITH HAPTEN 5-(PARA-NITROPHENYL PHOSPHONATE)-PENTANOIC ACID. AFFINITY MATURATION OF AN ESTEROLYTIC ANTIBODY 1ao7 
COMPLEX BETWEEN HUMAN T-CELL RECEPTOR, VIRAL PEPTIDE (TAX), AND HLA-A 0201 1aqk 
THREE-DIMENSIONAL STRUCTURE OF A HUMAN FAB WITH HIGH AFFINITY FOR TETANUS TOXOID 1axs 
MATURE OXY-COPE CATALYTIC ANTIBODY WITH HAPTEN 1axt 
IMMUNE VERSUS NATURAL SELECTION: ANTIBODY ALDOLASES WITH THE RATES OF NATURAL ENZYMES 1ay1 
ANTI TAQ FAB TP7 1b2w 
COMPARISON OF THE THREE-DIMENSIONAL STRUCTURES OF A HUMANIZED AND A CHIMERIC FAB OF AN ANTI-GAMMA-INTERFERON ANTIBODY 1b4j 
COMPARISON OF THE THREE-DIMENSIONAL STRUCTURES OF A HUMANIZED AND A CHIMERIC FAB OF AN ANTI-GAMMA-INTERFERON ANTIBODY 1b6d 
BENCE JONES PROTEIN DEL: AN ENTIRE IMMUNOGLOBULIN KAPPA LIGHT-CHAIN DIMER 1b6u 
CRYSTAL STRUCTURE OF THE HUMAN KILLER CELL INHIBITORY RECEPTOR (KIR2DL3) SPECIFIC FOR HLA-CW3 RELATED ALLELES 1baf 
2.9 ANGSTROMS RESOLUTION STRUCTURE OF AN ANTI-DINITROPHENYL-SPIN-LABEL MONOCLONAL ANTIBODY FAB FRAGMENT WITH BOUND HAPTEN 1bbd 
THREE DIMENSIONAL STRUCTURE OF THE FAB FRAGMENT OF A NEUTRALIZING ANTIBODY TO HUMAN RHINOVIRUS SEROTYPE 2 1bbj 
CRYSTAL STRUCTURE OF A CHIMERIC FAB' FRAGMENT OF AN ANTIBODY BINDING TUMOUR CELLS 1bd2 
COMPLEX BETWEEN HUMAN T-CELL RECEPTOR B7, VIRAL PEPTIDE (TAX) AND MHC CLASS I MOLECULE HLA-A 0201 1bec 
BETA CHAIN OF A T CELL ANTIGEN RECEPTOR 1bey 
ANTIBODY TO CAMPATH-1H HUMANIZED FAB 1bfo 
CAMPATH-1G IGG2B RAT MONOCLONAL FAB 1bgx 
TAQ POLYMERASE IN COMPLEX WITH TP7, AN INHIBITORY FAB 1bih 
CRYSTAL STRUCTURE OF THE INSECT IMMUNE PROTEIN HEMOLIN: A NEW DOMAIN ARRANGEMENT WITH IMPLICATIONS FOR HOMOPHILIC ADHESION 1bj1 
VASCULAR ENDOTHELIAL GROWTH FACTOR IN COMPLEX WITH A NEUTRALIZING ANTIBODY 1bjm 
LOC NAKS, A LAMBDA 1 TYPE LIGHT-CHAIN DIMER (BENCE-JONES PROTEIN) CRYSTALLIZED IN NAKSO4 1bln 
ANTI-P-GLYCOPROTEIN FAB MRK-16 1bm3 
IMMUNOGLOBULIN OPG2 FAB-PEPTIDE COMPLEX 1bog 
ANTI-P24 (HIV-1) FAB FRAGMENT CB41 COMPLEXED WITH AN EPITOPE-HOMOLOGOUS PEPTIDE 1bql 
STRUCTURE OF AN ANTI-HEL FAB FRAGMENT COMPLEXED WITH BOBWHITE QUAIL LYSOZYME 1bz7 
FAB FRAGMENT FROM MURINE ASCITES 1c12 
INSIGHT IN ODORANT PERCEPTION: THE CRYSTAL STRUCTURE AND BINDING CHARACTERISTICS OF ANTIBODY FRAGMENTS DIRECTED AGAINST THE MUSK ODORANT TRASEOLIDE 1c1e 
CRYSTAL STRUCTURE OF A DIELS-ALDERASE CATALYTIC ANTIBODY 1E9 IN COMPLEX WITH ITS HAPTEN 1c5b 
DECARBOXYLASE CATALYTIC ANTIBODY 21D8 UNLIGANDED FORM 1c5c 
DECARBOXYLASE CATALYTIC ANTIBODY 21D8-HAPTEN COMPLEX 1c5d 
THE CRYSTAL STRUCTURE OF THE FAB FRAGMENT OF A RAT MONOCLONAL ANTIBODY AGAINST THE MAIN IMMUNOGENIC REGION OF THE HUMAN MUSCLE ACETYLCHOLINE RECEPTOR 1cbv 
AN AUTOANTIBODY TO SINGLE-STRANDED DNA: COMPARISON OF THE THREE-DIMENSIONAL STRUCTURES OF THE UNLIGANDED FAB AND A DEOXYNUCLEOTIDE-FAB COMPLEX 1cdh 
STRUCTURES OF AN HIV AND MHC BINDING FRAGMENT FROM HUMAN CD4 AS REFINED IN TWO CRYSTAL LATTICES 1cdi 
STRUCTURES OF AN HIV AND MHC BINDING FRAGMENT FROM HUMAN CD4 AS REFINED IN TWO CRYSTAL LATTICES 1cdj 
STRUCTURE OF T-CELL SURFACE GLYCOPROTEIN CD4 1cdu 
STRUCTURE OF T-CELL SURFACE GLYCOPROTEIN CD4 MUTANT WITH PHE 43 REPLACED BY VAL 1cdy 
STRUCTURE OF T-CELL SURFACE GLYCOPROTEIN CD4 MUTANT WITH GLY 47 REPLACED BY SER 1ce1 
1.9A STRUCTURE OF THE THERAPEUTIC ANTIBODY CAMPATH-1H FAB IN COMPLEX WITH A SYNTHETIC PEPTIDE ANTIGEN 1cf8 
Convergence of catalytic antibody and terpene cyclase mechanisms: polyene cyclization directed by carbocation-pi interactions 1cfn 
ANTI-P24 (HIV-1) FAB FRAGMENT CB41 COMPLEXED WITH AN EPITOPE-RELATED PEPTIDE 1cfq 
ANTI-P24 (HIV-1) FAB FRAGMENT CB41 1cfs 
ANTI-P24 (HIV-1) FAB FRAGMENT CB41 COMPLEXED WITH AN EPITOPE-UNRELATED PEPTIDE 1cft 
ANTI-P24 (HIV-1) FAB FRAGMENT CB41 COMPLEXED WITH AN EPITOPE-UNRELATED D-PEPTIDE 1cgs 
LOCAL AND TRANSMITTED CONFORMATIONAL CHANGES ON COMPLEXATION OF AN ANTI-SWEETENER FAB 1cic 
IDIOTOPE-ANTI-IDIOTOPE FAB-FAB COMPLEX; D1.3-E225 1ck0 
ANTI-ANTI-IDIOTYPIC ANTIBODY AGAINST HUMAN ANGIOTENSIN II, UNLIGANDED FORM 1cl7 
ANTI HIV1 PROTEASE FAB 1clo 
ANTI-CARCINOEMBRYONIC ANTIGEN MONOCLONAL ANTIBODY A5B7 1cly 
IGG FAB (HUMAN IGG1, KAPPA) CHIMERIC FRAGMENT (CBR96) COMPLEXED WITH LEWIS Y NONOATE METHYL ESTER 1clz 
IGG FAB (IGG3, KAPPA) FRAGMENT (MBR96) COMPLEXED WITH LEWIS Y NONOATE METHYL ESTER 1cr9 
CRYSTAL STRUCTURE OF THE ANTI-PRION FAB 3F4 1cs6 
N-TERMINAL FRAGMENT OF AXONIN-1 FROM CHICKEN 1ct8 
CATALYTIC ANTIBODY 7C8 COMPLEX 1cu4 
CRYSTAL STRUCTURE OF THE ANTI-PRION FAB 3F4 IN COMPLEX WITH ITS PEPTIDE EPITOPE 1cz8 
VASCULAR ENDOTHELIAL GROWTH FACTOR IN COMPLEX WITH AN AFFINITY MATURED ANTIBODY 1d5b 
UNLIGANDED MATURE OXY-COPE CATALYTIC ANTIBODY 1d5i 
UNLIGANDED GERMLINE PRECURSOR OF AN OXY-COPE CATALYTIC ANTIBODY 1d6v 
CONFORMATION EFFECTS IN BIOLOGICAL CATALYSIS INTRODUCED BY OXY-COPE ANTIBODY MATURATION 1dba 
THREE-DIMENSIONAL STRUCTURE OF AN ANTI-STEROID FAB' AND PROGESTERONE-FAB' COMPLEX 1dbb 
THREE-DIMENSIONAL STRUCTURE OF AN ANTI-STEROID FAB' AND PROGESTERONE-FAB' COMPLEX 1dbj 
MOLECULAR BASIS OF CROSS-REACTIVITY AND THE LIMITS OF ANTIBODY-ANTIGEN COMPLEMENTARITY 1dbk 
MOLECULAR BASIS OF CROSS-REACTIVITY AND THE LIMITS OF ANTIBODY-ANTIGEN COMPLEMENTARITY 1dbm 
MOLECULAR BASIS OF CROSS-REACTIVITY AND THE LIMITS OF ANTIBODY-ANTIGEN COMPLEMENTARITY 1dcl 
MCG, A LAMBDA V TYPE LIGHT-CHAIN DIMER (BENCE-JONES PROTEIN), CRYSTALLIZED FROM AMMONIUM SULFATE 1dee 
CRYSTAL STRUCTURE AT 2.7A RESOLUTION OF A COMPLEX BETWEEN A STAPHYLOCOCCUS AUREUS DOMAIN AND A FAB FRAGMENT OF A HUMAN IGM ANTIBODY 1dfb 
STRUCTURE OF A HUMAN MONOCLONAL ANTIBODY FAB FRAGMENT AGAINST GP41 OF HUMAN IMMUNODEFICIENCY VIRUS TYPE I 1dn0 
STRUCTURE OF THE FAB FRAGMENT FROM A HUMAN IGM COLD AGGLUTININ 1dn2 
FC FRAGMENT OF HUMAN IGG1 IN COMPLEX WITH AN ENGINEERED 13 RESIDUE PEPTIDE DCAWHLGELVWCT-NH2 1dqd 
CRYSTAL STRUCTURE OF FAB HGR-2 F6, A COMPETITIVE ANTAGONIST OF THE GLUCAGON RECEPTOR 1dqj 
CRYSTAL STRUCTURE OF THE ANTI-LYSOZYME ANTIBODY HYHEL-63 COMPLEXED WITH HEN EGG WHITE LYSOZYME 1dqm 
CRYSTAL STRUCTURE OF ANTI-LYSOZYME ANTIBODY 1dqq 
CRYSTAL STRUCTURE OF ANTI-LYSOZYME ANTIBODY HYHEL-63 1dr9 
CRYSTAL STRUCTURE OF A SOLUBLE FORM OF B7-1 (CD80) 1e07 
Model of human carcinoembryonic antigen by homology modelling and curve-fitting to experimental solution scattering data 1e4k 
Crystal structure of soluble human IgG1 Fc fragment-Fc-gamma Receptor III complex 1e4w 
crossreactive binding of a circularized peptide to an anti-TGFalpha antibody Fab-fragment 1e4x 
crossreactive binding of a circularized peptide to an anti-TGFalpha antibody Fab-fragment 1e6j 
Crystal structure of HIV-1 capsid protein (p24) in complex with Fab13B5 1e6o 
Crystal structure of Fab13B5 against HIV-1 capsid protein p24 1eap 
CRYSTAL STRUCTURE OF A CATALYTIC ANTIBODY WITH A SERINE PROTEASE ACTIVE SITE 1efx 
STRUCTURE OF A COMPLEX BETWEEN THE HUMAN NATURAL KILLER CELL RECEPTOR KIR2DL2 AND A CLASS I MHC LIGAND HLA-CW3 1egj 
DOMAIN 4 OF THE BETA COMMON CHAIN IN COMPLEX WITH AN ANTIBODY 1ehl 
64M-2 ANTIBODY FAB COMPLEXED WITH D(5HT)(6-4)T 1ejo 
FAB FRAGMENT OF NEUTRALISING MONOCLONAL ANTIBODY 4C4 COMPLEXED WITH G-H LOOP FROM FMDV. 1emt 
FAB ANTIBODY FRAGMENT OF AN C60 ANTIFULLERENE ANTIBODY 1eo8 
INFLUENZA VIRUS HEMAGGLUTININ COMPLEXED WITH A NEUTRALIZING ANTIBODY 1etz 
THE THREE-DIMENSIONAL STRUCTURE OF AN ANTI-SWEETENER FAB, NC10.14, SHOWS THE EXTENT OF STRUCTURAL DIVERSITY IN ANTIGEN RECOGNITION BY IMMUNOGLOBULINS 1f11 
F124 FAB FRAGMENT FROM A MONOCLONAL ANTI-PRES2 ANTIBODY 1f2q 
CRYSTAL STRUCTURE OF THE HUMAN HIGH-AFFINITY IGE RECEPTOR 1f3d 
CATALYTIC ANTIBODY 4B2 IN COMPLEX WITH ITS AMIDINIUM HAPTEN. 1f4w 
CRYSTAL STRUCTURE OF AN ANTI-CARBOHYDRATE ANTIBODY DIRECTED AGAINST VIBRIO CHOLERAE O1 IN COMPLEX WITH ANTIGEN 1f4x 
CRYSTAL STRUCTURE OF AN ANTI-CARBOHYDRATE ANTIBODY DIRECTED AGAINST VIBRIO CHOLERAE O1 IN COMPLEX WITH ANTIGEN 1f4y 
CRYSTAL STRUCTURE OF AN ANTI-CARBOHYDRATE ANTIBODY DIRECTED AGAINST VIBRIO CHOLERAE O1 IN COMPLEX WITH ANTIGEN 1f58 
IGG1 FAB FRAGMENT (58.2) COMPLEX WITH 24-RESIDUE PEPTIDE (RESIDUES 308-333 OF HIV-1 GP120 (MN ISOLATE) WITH ALA TO AIB SUBSTITUTION AT POSITION 323 1f6a 
Structure of the human ige-fc bound to its high affinity receptor fc(epsilon)ri(alpha) 1f8t 
FAB (LNKB-2) OF MONOCLONAL ANTIBODY, CRYSTAL STRUCTURE 1f90 
FAB FRAGMENT OF MONOCLONAL ANTIBODY (LNKB-2) AGAINST HUMAN INTERLEUKIN-2 IN COMPLEX WITH ANTIGENIC PEPTIDE 1f97 
SOLUBLE PART OF THE JUNCTION ADHESION MOLECULE FROM MOUSE 1fai 
THREE-DIMENSIONAL STRUCTURE OF TWO CRYSTAL FORMS OF FAB R19.9, FROM A MONOCLONAL ANTI-ARSONATE ANTIBODY 1fbi 
CRYSTAL STRUCTURE OF A CROSS-REACTION COMPLEX BETWEEN FAB F9.13.7 AND GUINEA-FOWL LYSOZYME 1fc1 
CRYSTALLOGRAPHIC REFINEMENT AND ATOMIC MODELS OF A HUMAN FC FRAGMENT AND ITS COMPLEX WITH FRAGMENT B OF PROTEIN A FROM STAPHYLOCOCCUS AUREUS AT 2.9-AND 2.8-ANGSTROMS RESOLUTION 1fc2 
CRYSTALLOGRAPHIC REFINEMENT AND ATOMIC MODELS OF A HUMAN FC FRAGMENT AND ITS COMPLEX WITH FRAGMENT B OF PROTEIN A FROM STAPHYLOCOCCUS AUREUS AT 2.9-AND 2.8-ANGSTROMS RESOLUTION 1fcc 
CRYSTAL STRUCTURE OF THE C2 FRAGMENT OF STREPTOCOCCAL PROTEIN G IN COMPLEX WITH THE FC DOMAIN OF HUMAN IGG 1fdl 
CRYSTALLOGRAPHIC REFINEMENT OF THE THREE-DIMENSIONAL STRUCTURE OF THE FAB D1.3-LYSOZYME COMPLEX AT 2.5-ANGSTROMS RESOLUTION 1fe8 
CRYSTAL STRUCTURE OF THE VON WILLEBRAND FACTOR A3 DOMAIN IN COMPLEX WITH A FAB FRAGMENT OF IGG RU5 THAT INHIBITS COLLAGEN BINDING 1fgn 
MONOCLONAL MURINE ANTIBODY 5G9-ANTI-HUMAN TISSUE FACTOR 1fh5 
CRYSTAL STRUCTURE OF THE FAB FRAGMENT OF THE MONOCLONAL ANTIBODY MAK33 1fig 
ROUTES TO CATALYSIS: STRUCTURE OF A CATALYTIC ANTIBODY AND COMPARISON WITH ITS NATURAL COUNTERPART 1fj1 
LYME DISEASE ANTIGEN OSPA IN COMPLEX WITH NEUTRALIZING ANTIBODY FAB LA-2 1fl3 
CRYSTAL STRUCTURE OF THE BLUE FLUORESCENT ANTIBODY (19G2) IN COMPLEX WITH STILBENE HAPTEN AT 277K 1fl5 
THE UNLIGANDED GERMLINE PRECURSOR TO THE SULFIDE OXIDASE CATALYTIC ANTIBODY 28B4. 1fl6 
THE HAPTEN COMPLEXED GERMLINE PRECURSOR TO SULFIDE OXIDASE CATALYTIC ANTIBODY 28B4 1flr 
4-4-20 FAB FRAGMENT 1fn4 
CRYSTAL STRUCTURE OF FAB198, AN EFFICIENT PROTECTOR OF ACETYLCHOLINE RECEPTOR AGAINST MYASTHENOGENIC ANTIBODIES 1fns 
CRYSTAL STRUCTURE OF THE VON WILLEBRAND FACTOR (VWF) A1 DOMAIN I546V MUTANT IN COMPLEX WITH THE FUNCTION BLOCKING FAB NMC4 1for 
STRUCTURE DETERMINATION OF AN FAB FRAGMENT THAT NEUTRALIZES HUMAN RHINOVIRUS AND ANALYSIS OF THE FAB-VIRUS COMPLEX 1fpt 
THREE-DIMENSIONAL STRUCTURE OF THE COMPLEX BETWEEN THE FAB FRAGMENT OF AN NEUTRALIZING ANTIBODY FOR TYPE 1 POLIOVIRUS AND ITS VIRAL EPITOPE 1frg 
CRYSTAL STRUCTURE, SEQUENCE, AND EPITOPE MAPPING OF A PEPTIDE COMPLEX OF AN ANTI-INFLUENZA HA PEPTIDE ANTIBODY FAB 26(SLASH)9: FINE-TUNING ANTIBODY SPECIFICITY 1frt 
CRYSTAL STRUCTURE OF THE COMPLEX OF RAT NEONATAL FC RECEPTOR WITH FC 1fsk 
COMPLEX FORMATION BETWEEN A FAB FRAGMENT OF A MONOCLONAL IGG ANTIBODY AND THE MAJOR ALLERGEN FROM BIRCH POLLEN BET V 1 1fvd 
X-RAY STRUCTURES OF THE ANTIGEN-BINDING DOMAINS FROM THREE VARIANTS OF HUMANIZED ANTI-P185-HER2 ANTIBODY 4D5 AND COMPARISON WITH MOLECULAR MODELING 1fve 
X-RAY STRUCTURES OF THE ANTIGEN-BINDING DOMAINS FROM THREE VARIANTS OF HUMANIZED ANTI-P185-HER2 ANTIBODY 4D5 AND COMPARISON WITH MOLECULAR MODELING 1fyt 
CRYSTAL STRUCTURE OF A COMPLEX OF A HUMAN ALPHA/BETA-T CELL RECEPTOR, INFLUENZA HA ANTIGEN PEPTIDE, AND MHC CLASS II MOLECULE, HLA-DR1 1g0x 
CRYSTAL STRUCTURE OF THE LIGAND BINDING DOMAIN OF LIR-1 (ILT2) 1g0y 
IL-1 RECEPTOR TYPE 1 COMPLEXED WITH ANTAGONIST PEPTIDE AF10847 1g6r 
A FUNCTIONAL HOT SPOT FOR ANTIGEN RECOGNITION IN A SUPERAGONIST TCR/MHC COMPLEX 1g9m 
HIV-1 HXBC2 GP120 ENVELOPE GLYCOPROTEIN COMPLEXED WITH CD4 AND INDUCED NEUTRALIZING ANTIBODY 17B 1g9n 
HIV-1 YU2 GP120 ENVELOPE GLYCOPROTEIN COMPLEXED WITH CD4 AND INDUCED NEUTRALIZING ANTIBODY 17B 1gaf 
48G7 HYBRIDOMA LINE FAB COMPLEXED WITH HAPTEN 5-(PARA-NITROPHENYL PHOSPHONATE)-PENTANOIC ACID 1gc1 
HIV-1 GP120 CORE COMPLEXED WITH CD4 AND A NEUTRALIZING HUMAN ANTIBODY 1ggb 
MAJOR ANTIGEN-INDUCED DOMAIN REARRANGEMENTS IN AN ANTIBODY 1ggc 
MAJOR ANTIGEN-INDUCED DOMAIN REARRANGEMENTS IN AN ANTIBODY 1ggi 
CRYSTAL STRUCTURE OF AN HIV-1 NEUTRALIZING ANTIBODY 50.1 IN COMPLEX WITH ITS V3 LOOP PEPTIDE ANTIGEN 1ghf 
ANTI-ANTI-IDIOTYPE GH1002 FAB FRAGMENT 1gig 
REFINED THREE-DIMENSIONAL STRUCTURE OF THE FAB FRAGMENT OF A MURINE IGG1, LAMBDA ANTIBODY 1gpo 
CRYSTAL STRUCTURE OF THE RATIONALLY DESIGNED ANTIBODY M41 AS A FAB FRAGMENT 1h0d 
Crystal structure of Human Angiogenin in complex with Fab fragment of its monoclonal antibody mAb 26-2F 1h3p 
STRUCTURAL CHARACTERISATION OF A MONOCLONAL ANTIBODY SPECIFIC FOR THE PRES1 REGION OF THE HEPATITIS B VIRUS 1h3t 
Crystal structure of the human igg1 fc-fragment,glycoform (mn2f)2 1h3u 
Structural analysis of human IgG-Fc glycoforms reveals a correlation between glycosylation and structural integrity 1h3v 
Structural analysis of human IgG-Fc glycoforms reveals a correlation between glycosylation and structural integrity 1h3w 
Structural analysis of human IgG-Fc glycoforms reveals a correlation between glycosylation and structural integrity 1h3x 
Structural analysis of human IgG-Fc glycoforms reveals a correlation between glycosylation and structural integrity 1h3y 
Structural analysis of human IgG-Fc glycoforms reveals a correlation between glycosylation and structural integrity 1hez 
antibody-antigen complex 1hh6 
ANTI-P24 (HIV-1) FAB FRAGMENT CB41 COMPLEXED WITH A PEPTIDE 1hh9 
ANTI-P24 (HIV-1) FAB FRAGMENT CB41 COMPLEXED WITH A PEPTIDE 1hi6 
ANTI-P24 (HIV-1) FAB FRAGMENT CB41 COMPLEXED WITH A PEPTIDE 1hil 
STRUCTURAL EVIDENCE FOR INDUCED FIT AS A MECHANISM FOR ANTIGEN-ANTIBODY RECOGNITION 1him 
STRUCTURAL EVIDENCE FOR INDUCED FIT AS A MECHANISM FOR ANTIBODY-ANTIGEN RECOGNITION 1hin 
STRUCTURAL EVIDENCE FOR INDUCED FIT AS A MECHANISM FOR ANTIBODY-ANTIGEN RECOGNITION 1hkl 
FREE AND LIGANDED FORM OF AN ESTEROLYTIC CATALYTIC ANTIBODY 1hq4 
STRUCTURE OF NATIVE CATALYTIC ANTIBODY HA5-19A4 1hxm 
Crystal Structure of a Human Vgamma9/Vdelta2 T Cell Receptor 1hys 
CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH A POLYPURINE TRACT RNA:DNA 1hzh 
CRYSTAL STRUCTURE OF THE INTACT HUMAN IGG B12 WITH BROAD AND POTENT ACTIVITY AGAINST PRIMARY HIV-1 ISOLATES: A TEMPLATE FOR HIV VACCINE DESIGN 1i7z 
ANTIBODY GNC92H2 BOUND TO LIGAND 1i8l 
HUMAN B7-1/CTLA-4 CO-STIMULATORY COMPLEX 1i8m 
CRYSTAL STRUCTURE OF A RECOMBINANT ANTI-SINGLE-STRANDED DNA ANTIBODY FRAGMENT COMPLEXED WITH DT5 1i9i 
NATIVE CRYSTAL STRUCTURE OF THE RECOMBINANT MONOCLONAL WILD TYPE ANTI-TESTOSTERONE FAB FRAGMENT 1i9j 
TESTOSTERONE COMPLEX STRUCTURE OF THE RECOMBINANT MONOCLONAL WILD TYPE ANTI-TESTOSTERONE FAB FRAGMENT 1i9r 
STRUCTURE OF CD40L IN COMPLEX WITH THE FAB FRAGMENT OF HUMANIZED 5C8 ANTIBODY 1iai 
IDIOTYPE-ANTI-IDIOTYPE FAB COMPLEX 1ibg 
STRUCTURE AND SPECIFICITY OF THE ANTI-DIGOXIN ANTIBODY 40-50 1ifh 
A DETAILED ANALYSIS OF THE FREE AND BOUND CONFORMATION OF AN ANTIBODY: X-RAY STRUCTURES OF ANTI-PEPTIDE FAB 17(SLASH)9 AND THREE DIFFERENT FAB-PEPTIDE COMPLEXES 1iga 
MODEL OF HUMAN IGA1 DETERMINED BY SOLUTION SCATTERING CURVE-FITTING AND HOMOLOGY MODELLING 1igc 
IGG1 FAB FRAGMENT (MOPC21) COMPLEX WITH DOMAIN III OF PROTEIN G FROM STREPTOCOCCUS 1igf 
CRYSTAL STRUCTURES OF AN ANTIBODY TO A PEPTIDE AND ITS COMPLEX WITH PEPTIDE ANTIGEN AT 2.8 ANGSTROMS 1igi 
26-10 FAB:DIGOXIN COMPLEX-AFFINITY AND SPECIFICITY DUE TO SURFACE COMPLEMENTARITY 1igj 
26-10 FAB:DIGOXIN COMPLEX-AFFINITY AND SPECIFICITY DUE TO SURFACE COMPLEMENTARITY 1igt 
STRUCTURE OF IMMUNOGLOBULIN 1igy 
STRUCTURE OF IMMUNOGLOBULIN 1ikf 
A CONFORMATION OF CYCLOSPORIN A IN AQUEOUS ENVIRONMENT REVEALED BY THE X-RAY STRUCTURE OF A CYCLOSPORIN-FAB COMPLEX 1il1 
Crystal structure of G3-519, an anti-HIV monoclonal antibody 1im9 
Crystal structure of the human natural killer cell inhibitory receptor KIR2DL1 bound to its MHC ligand HLA-Cw4 1ind 
HOW THE ANTI-(METAL CHELATE) ANTIBODY CHA255 IS SPECIFIC FOR THE METAL ION OF ITS ANTIGEN: X-RAY STRUCTURES FOR TWO FAB'(SLASH)HAPTEN COMPLEXES WITH DIFFERENT METALS IN THE CHELATE 1ine 
HOW THE ANTI-(METAL CHELATE) ANTIBODY CHA255 IS SPECIFIC FOR THE METAL ION OF ITS ANTIGEN: X-RAY STRUCTURES FOR TWO FAB'(SLASH)HAPTEN COMPLEXES WITH DIFFERENT METALS IN THE CHELATE 1iqd 
Human Factor VIII C2 Domain complexed to human monoclonal BO2C11 Fab. 1iqw 
CRYSTAL STRUCTURE OF THE FAB FRAGMENT OF THE MOUSE ANTI-HUMAN FAS ANTIBODY HFE7A 1ira 
COMPLEX OF THE INTERLEUKIN-1 RECEPTOR WITH THE INTERLEUKIN-1 RECEPTOR ANTAGONIST (IL1RA) 1it9 
CRYSTAL STRUCTURE OF AN ANTIGEN-BINDING FRAGMENT FROM A HUMANIZED VERSION OF THE ANTI-HUMAN FAS ANTIBODY HFE7A 1itb 
TYPE-1 INTERLEUKIN-1 RECEPTOR COMPLEXED WITH INTERLEUKIN-1 BETA 1j5o 
CRYSTAL STRUCTURE OF MET184ILE MUTANT OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH DOUBLE STRANDED DNA TEMPLATE-PRIMER 1j86 
HUMAN HIGH AFFINITY FC RECEPTOR FC(EPSILON)RI(ALPHA), MONOCLINIC CRYSTAL FORM 2 1j87 
HUMAN HIGH AFFINITY FC RECEPTOR FC(EPSILON)RI(ALPHA), HEXAGONAL CRYSTAL FORM 1 1j88 
HUMAN HIGH AFFINITY FC RECEPTOR FC(EPSILON)RI(ALPHA), TETRAGONAL CRYSTAL FORM 1 1j89 
HUMAN HIGH AFFINITY FC RECEPTOR FC(EPSILON)RI(ALPHA), TETRAGONAL CRYSTAL FORM 2 1j8h 
Crystal Structure of a Complex of a Human alpha/beta-T cell Receptor, Influenza HA Antigen Peptide, and MHC Class II Molecule, HLA-DR4 1jck 
T-CELL RECEPTOR BETA CHAIN COMPLEXED WITH SEC3 SUPERANTIGEN 1jfq 
ANTIGEN-BINDING FRAGMENT OF THE MURINE ANTI-PHENYLARSONATE ANTIBODY 36-71, ""FAB 36-71"" 1jgl 
Crystal structure of immunoglobulin Fab fragment complexed with 17-beta-estradiol 1jgu 
STRUCTURAL BASIS FOR DISFAVORED ELIMINATION REACTION IN CATALYTIC ANTIBODY 1D4 1jgv 
STRUCTURAL BASIS FOR DISFAVORED ELIMINATION REACTION IN CATALYTIC ANTIBODY 1D4 1jhk 
Crystal structure of the anti-estradiol antibody 57-2 1jl4 
CRYSTAL STRUCTURE OF THE HUMAN CD4 N-TERMINAL TWO DOMAIN FRAGMENT COMPLEXED TO A CLASS II MHC MOLECULE 1jn6 
Crystal Structure of Fab-Estradiol Complexes 1jnh 
Crystal Structure of Fab-Estradiol Complexes 1jnl 
Crystal Structure of Fab-Estradiol Complexes 1jnn 
Crystal Structure of Fab-Estradiol Complexes 1jps 
Crystal structure of tissue factor in complex with humanized Fab D3h44 1jpt 
Crystal Structure of Fab D3H44 1jrh 
COMPLEX (ANTIBODY/ANTIGEN) 1jvk 
THREE-DIMENSIONAL STRUCTURE OF AN IMMUNOGLOBULIN LIGHT CHAIN DIMER ACTING AS A LETHAL AMYLOID PRECURSOR 1k4c 
Potassium Channel KcsA-Fab complex in high concentration of K+ 1k4d 
Potassium Channel KcsA-Fab complex in low concentration of K+ 1k6q 
Crystal structure of antibody Fab fragment D3 1kb5 
MURINE T-CELL RECEPTOR VARIABLE DOMAIN/FAB COMPLEX 1kc5 
CRYSTAL STRUCTURE OF ANTIBODY PC287 IN COMPLEX WITH PS1 PEPTIDE 1kcr 
CRYSTAL STRUCTURE OF ANTIBODY PC283 IN COMPLEX WITH PS1 PEPTIDE 1kcs 
CRYSTAL STRUCTURE OF ANTIBODY PC282 IN COMPLEX WITH PS1 PEPTIDE 1kcu 
CRYSTAL STRUCTURE OF ANTIBODY PC287 1kcv 
Crystal structure of antibody pc282 1keg 
Antibody 64M-2 Fab complexed with dTT(6-4)TT 1kel 
CATALYTIC ANTIBODY 28B4 FAB FRAGMENT COMPLEXED WITH HAPTEN (1-[N-4'-NITROBENZYL-N-4'-CARBOXYBUTYLAMINO] METHYLPHOSPHONIC ACID) 1kem 
CATALYTIC ANTIBODY 28B4 FAB FRAGMENT 1ken 
INFLUENZA VIRUS HEMAGGLUTININ COMPLEXED WITH AN ANTIBODY THAT PREVENTS THE HEMAGGLUTININ LOW PH FUSOGENIC TRANSITION 1kfa 
Crystal structure of Fab fragment complexed with gibberellin A4 1kgc 
Immune Receptor 1kn2 
CATALYTIC ANTIBODY D2.3 COMPLEX 1kn4 
CATALYTIC ANTIBODY D2.3 COMPLEX 1kno 
CRYSTAL STRUCTURE OF THE COMPLEX OF A CATALYTIC ANTIBODY FAB WITH A TRANSITION STATE ANALOG: STRUCTURAL SIMILARITIES IN ESTERASE-LIKE ABZYMES 1ktk 
Complex of Streptococcal pyrogenic enterotoxin C (SpeC) with a human T cell receptor beta chain (Vbeta2.1) 1l0x 
TCR beta chain complexed with streptococcal superantigen SpeA 1l0y 
T cell receptor beta chain complexed with superantigen SpeA soaked with zinc 1l6x 
FC FRAGMENT OF RITUXIMAB BOUND TO A MINIMIZED VERSION OF THE B-DOMAIN FROM PROTEIN A CALLED Z34C 1l7i 
Crystal Structure of the anti-ErbB2 Fab2C4 1l7t 
Crystal Structure Analysis of the anti-testosterone Fab fragment 1lgv 
Structure of a Human Bence-Jones Dimer Crystallized in U.S. Space Shuttle Mission STS-95: 100K 1lhz 
Structure of a Human Bence-Jones Dimer Crystallized in U.S. Space Shuttle Mission STS-95: 293K 1lil 
BENCE JONES PROTEIN CLE, A LAMBDA III IMMUNOGLOBULIN LIGHT-CHAIN DIMER 1lk3 
ENGINEERED HUMAN INTERLEUKIN-10 MONOMER COMPLEXED TO 9D7 FAB FRAGMENT 1lo0 
Catalytic Retro-Diels-Alderase Transition State Analogue Complex 1lo2 
Retro-Diels-Alderase Catalytic Antibody 1lo3 
Retro-Diels-Alderase Catalytic Antibody: Product Analogue 1lo4 
Retro-Diels-Alderase Catalytic antibody 9D9 1lp9 
Xenoreactive complex AHIII 12.2 TCR bound to p1049/HLA-A2.1 1m4k 
Crystal structure of the human natural killer cell activator receptor KIR2DS2 (CD158j) 1m71 
Crystal structure of a Monoclonal Fab Specific for Shigella Flexneri Y lipopolysaccharide 1m7d 
Crystal structure of a Monoclonal Fab Specific for Shigella flexneri Y Lipopolysaccharide complexed with a trisaccharide 1m7i 
CRYSTAL STRUCTURE OF A MONOCLONAL FAB SPECIFIC FOR SHIGELLA FLEXNERI Y LIPOPOLYSACCHARIDE COMPLEXED WITH A PENTASACCHARIDE 1mam 
CRYSTAL STRUCTURE TO 2.45 A RESOLUTION OF A MONOCLONAL FAB SPECIFIC FOR THE BRUCELLA A CELL WALL POLYSACCHARIDE ANTIGEN 1mcb 
PRINCIPLES AND PITFALLS IN DESIGNING SITE DIRECTED PEPTIDE LIGANDS 1mcc 
PRINCIPLES AND PITFALLS IN DESIGNING SITE DIRECTED PEPTIDE LIGANDS 1mcd 
PRINCIPLES AND PITFALLS IN DESIGNING SITE DIRECTED PEPTIDE LIGANDS 1mce 
PRINCIPLES AND PITFALLS IN DESIGNING SITE DIRECTED PEPTIDE LIGANDS 1mcf 
PRINCIPLES AND PITFALLS IN DESIGNING SITE DIRECTED PEPTIDE LIGANDS 1mch 
PRINCIPLES AND PITFALLS IN DESIGNING SITE DIRECTED PEPTIDE LIGANDS 1mci 
PRINCIPLES AND PITFALLS IN DESIGNING SITE DIRECTED PEPTIDE LIGANDS 1mcj 
PRINCIPLES AND PITFALLS IN DESIGNING SITE DIRECTED PEPTIDE LIGANDS 1mck 
PRINCIPLES AND PITFALLS IN DESIGNING SITE DIRECTED PEPTIDE LIGANDS 1mcl 
PRINCIPLES AND PITFALLS IN DESIGNING SITE DIRECTED PEPTIDE LIGANDS 1mcn 
PRINCIPLES AND PITFALLS IN DESIGNING SITE DIRECTED PEPTIDE LIGANDS 1mco 
THREE-DIMENSIONAL STRUCTURE OF A HUMAN IMMUNOGLOBULIN WITH A HINGE DELETION 1mcp 
PHOSPHOCHOLINE BINDING IMMUNOGLOBULIN FAB MC/PC603. AN X-RAY DIFFRACTION STUDY AT 2.7 ANGSTROMS 1mcq 
PRINCIPLES AND PITFALLS IN DESIGNING SITE DIRECTED PEPTIDE LIGANDS 1mcr 
PRINCIPLES AND PITFALLS IN DESIGNING SITE DIRECTED PEPTIDE LIGANDS 1mcs 
PRINCIPLES AND PITFALLS IN DESIGNING SITE DIRECTED PEPTIDE LIGANDS 1mcw 
THREE-DIMENSIONAL STRUCTURE OF A HYBRID LIGHT CHAIN DIMER. PROTEIN ENGINEERING OF A BINDING CAVITY 1mex 
Antibody Catalysis of a Bimolecular Cycloaddition Reaction 1mf2 
ANTI HIV1 PROTEASE FAB COMPLEX 1mfb 
HIGH RESOLUTION STRUCTURES OF ANTIBODY FAB FRAGMENT COMPLEXED WITH CELL-SURFACE OLIGOSACCHARIDE OF PATHOGENIC SALMONELLA 1mfc 
HIGH RESOLUTION STRUCTURES OF ANTIBODY FAB FRAGMENT COMPLEXED WITH CELL-SURFACE OLIGOSACCHARIDE OF PATHOGENIC SALMONELLA 1mfd 
THE SOLUTION STRUCTURE OF A TRISACCHARIDE-ANTIBODY COMPLEX: COMPARISON OF NMR MEASUREMENTS WITH A CRYSTAL STRUCTURE 1mfe 
RECOGNITION OF A CELL-SURFACE OLIGO-SACCHARIDE OF PATHOGENIC SALMONELLA BY AN ANTIBODY FAB FRAGMENT 1mh5 
The Structure Of The Complex Of The Fab Fragment Of The Esterolytic Antibody MS6-164 and A Transition-State Analog 1mhh 
Antibody-antigen complex 1mhp 
Crystal structure of a chimeric alpha1 integrin I-domain in complex with the Fab fragment of a humanized neutralizing antibody 1mi5 
The crystal structure of LC13 TcR in complex with HLAB8-EBV peptide complex 1mie 
Crystal Structure Of The Fab Fragment of Esterolytic Antibody MS5-393 1mim 
IGG FAB FRAGMENT (CD25-BINDING) 1mj7 
Crystal Structure Of The Complex Of The Fab fragment of Esterolytic Antibody MS5-393 and A Transition-State Analog 1mj8 
High Resolution Crystal Structure Of The Fab Fragment of The Esterolytic Antibody MS6-126 1mjj 
HIGH RESOLUTION CRYSTAL STRUCTURE OF THE COMPLEX OF THE FAB FRAGMENT OF ESTEROLYTIC ANTIBODY MS6-12 AND A TRANSITION-STATE ANALOG 1mju 
1.22 ANGSTROM RESOLUTION CRYSTAL STRUCTURE OF THE FAB FRAGMENT OF ESTEROLYTIC ANTIBODY MS6-12 1mlb 
MONOCLONAL ANTIBODY FAB D44.1 RAISED AGAINST CHICKEN EGG-WHITE LYSOZYME 1mlc 
MONOCLONAL ANTIBODY FAB D44.1 RAISED AGAINST CHICKEN EGG-WHITE LYSOZYME COMPLEXED WITH LYSOZYME 1mnu 
UNLIGANDED BACTERICIDAL ANTIBODY AGAINST NEISSERIA MENINGITIDIS 1mpa 
BACTERICIDAL ANTIBODY AGAINST NEISSERIA MENINGITIDIS 1mq8 
Crystal structure of alphaL I domain in complex with ICAM-1 1mrc 
PREPARATION, CHARACTERIZATION AND CRYSTALLIZATION OF AN ANTIBODY FAB FRAGMENT THAT RECOGNIZES RNA. CRYSTAL STRUCTURES OF NATIVE FAB AND THREE FAB-MONONUCLEOTIDE COMPLEXES 1mrd 
PREPARATION, CHARACTERIZATION AND CRYSTALLIZATION OF AN ANTIBODY FAB FRAGMENT THAT RECOGNIZES RNA. CRYSTAL STRUCTURES OF NATIVE FAB AND THREE FAB-MONONUCLEOTIDE COMPLEXES 1mre 
PREPARATION, CHARACTERIZATION AND CRYSTALLIZATION OF AN ANTIBODY FAB FRAGMENT THAT RECOGNIZES RNA. CRYSTAL STRUCTURES OF NATIVE FAB AND THREE FAB-MONONUCLEOTIDE COMPLEXES 1mrf 
PREPARATION, CHARACTERIZATION AND CRYSTALLIZATION OF AN ANTIBODY FAB FRAGMENT THAT RECOGNIZES RNA. CRYSTAL STRUCTURES OF NATIVE FAB AND THREE FAB-MONONUCLEOTIDE COMPLEXES 1mwa 
2C/H-2KBM3/DEV8 ALLOGENEIC COMPLEX 1n0x 
Crystal Structure of a Broadly Neutralizing Anti-HIV-1 Antibody in Complex with a Peptide Mimotope 1n5y 
HIV-1 Reverse Transcriptase Crosslinked to Post-Translocation AZTMP-Terminated DNA (Complex P) 1n64 
Crystal structure analysis of the immunodominant antigenic site on Hepatitis C virus protein bound to mAb 19D9D6 1n6q 
HIV-1 Reverse Transcriptase Crosslinked to pre-translocation AZTMP-terminated DNA (complex N) 1n7m 
Germline 7G12 with N-methylmesoporphyrin 1n8z 
Crystal structure of extracellular domain of human HER2 complexed with Herceptin Fab 1nak 
IGG1 FAB FRAGMENT (83.1) COMPLEX WITH 16-RESIDUE PEPTIDE (RESIDUES 304-321 OF HIV-1 GP120 (MN ISOLATE)) 1nbq 
Crystal Structure of Human Junctional Adhesion Molecule Type 1 1nbv 
AN AUTOANTIBODY TO SINGLE-STRANDED DNA: COMPARISON OF THE THREE-DIMENSIONAL STRUCTURES OF THE UNLIGANDED FAB AND A DEOXYNUCLEOTIDE-FAB COMPLEX 1nby 
Crystal Structure of HyHEL-63 complexed with HEL mutant K96A 1nbz 
Crystal Structure of HyHEL-63 complexed with HEL mutant K97A 1nc2 
Crystal Structure of Monoclonal Antibody 2D12.5 Fab Complexed with Y-DOTA 1nc4 
Crystal Structure of Monoclonal Antibody 2D12.5 Fab Complexed with Gd-DOTA 1nca 
REFINED CRYSTAL STRUCTURE OF THE INFLUENZA VIRUS N9 NEURAMINIDASE-NC41 FAB COMPLEX 1ncb 
CRYSTAL STRUCTURES OF TWO MUTANT NEURAMINIDASE-ANTIBODY COMPLEXES WITH AMINO ACID SUBSTITUTIONS IN THE INTERFACE 1ncc 
CRYSTAL STRUCTURES OF TWO MUTANT NEURAMINIDASE-ANTIBODY COMPLEXES WITH AMINO ACID SUBSTITUTIONS IN THE INTERFACE 1ncd 
REFINED CRYSTAL STRUCTURE OF THE INFLUENZA VIRUS N9 NEURAMINIDASE-NC41 FAB COMPLEX 1ncw 
Cationic Cyclization Antibody 4C6 in Complex with Benzoic Acid 1nd0 
CATIONIC CYCLIZATION ANTIBODY 4C6 COMPLEX WITH TRANSITION STATE ANALOG 1ndg 
Crystal structure of Fab fragment of antibody HyHEL-8 complexed with its antigen lysozyme 1ndm 
Crystal structure of Fab fragment of antibody HyHEL-26 complexed with lysozyme 1nfd 
AN ALPHA-BETA T CELL RECEPTOR (TCR) HETERODIMER IN COMPLEX WITH AN ANTI-TCR FAB FRAGMENT DERIVED FROM A MITOGENIC ANTIBODY 1ngp 
N1G9 (IGG1-LAMBDA) FAB FRAGMENT COMPLEXED WITH (4-HYDROXY-3-NITROPHENYL) ACETATE 1ngq 
N1G9 (IGG1-LAMBDA) FAB FRAGMENT 1ngw 
Chimeric Affinity Matured Fab 7g12 complexed with mesoporphyrin 1ngx 
Chimeric Germline Fab 7g12 with jeffamine fragment bound 1ngy 
Chimeric Mature Fab 7g12-Apo 1ngz 
Chimeric Germline Fab 7g12-apo 1nj9 
Cocaine hydrolytic antibody 15A10 1nkr 
INHIBITORY RECEPTOR (P58-CL42) FOR HUMAN NATURAL KILLER CELLS 1nl0 
Crystal structure of human factor IX Gla domain in complex of an inhibitory antibody, 10C12 1nlb 
crystal structure of anti-HCV monoclonal antibody 19D9D6 1nld 
FAB FRAGMENT OF A NEUTRALIZING ANTIBODY DIRECTED AGAINST AN EPITOPE OF GP41 FROM HIV-1 1nsn 
THE CRYSTAL STRUCTURE OF ANTIBODY N10-STAPHYLOCOCCAL NUCLEASE COMPLEX AT 2.9 ANGSTROMS RESOLUTION 1oak 
CRYSTAL STRUCTURE OF THE VON WILLEBRAND FACTOR (VWF) A1 DOMAIN IN COMPLEX WITH THE FUNCTION BLOCKING NMC-4 FAB 1ob1 
Crystal structure of a Fab complex whith Plasmodium falciparum MSP1-19 1oga 
A structural basis for immunodominant human T-cell receptor recognition. 1oll 
Extracellular region of the human receptor NKp46 1om3 
FAB 2G12 unliganded 1op3 
Crystal Structure of Fab 2G12 bound to Man1->2Man 1op5 
Crystal Structure of Fab 2G12 bound to Man9GlcNAc2 1opg 
OPG2 FAB FRAGMENT 1oqo 
Complex between G0 version of an Fc bound to a minimized version of Protein A called Mini-Z 1oqx 
G-2 glycovariant of human IgG Fc bound to minimized version of Protein A called Z34C 1orq 
X-ray structure of a voltage-dependent potassium channel in complex with an Fab 1ors 
X-ray structure of the KvAP potassium channel voltage sensor in complex with an Fab 1osp 
CRYSTAL STRUCTURE OF OUTER SURFACE PROTEIN A OF BORRELIA BURGDORFERI COMPLEXED WITH A MURINE MONOCLONAL ANTIBODY FAB 1ots 
Structure of the Escherichia coli ClC Chloride channel and Fab Complex 1ott 
Structure of the Escherichia coli ClC Chloride channel E148A mutant and Fab Complex 1otu 
Structure of the Escherichia coli ClC Chloride channel E148Q mutant and Fab Complex 1p2c 
crystal structure analysis of an anti-lysozyme antibody 1p53 
The Crystal Structure of ICAM-1 D3-D5 fragment 1p6f 
Structure of the human natural cytotoxicity receptor NKp46 1p7k 
Crystal structure of an anti-ssDNA antigen-binding fragment (Fab) bound to 4-(2-Hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES) 1p7q 
Crystal Structure of HLA-A2 Bound to LIR-1, a Host and Viral MHC Receptor 1pg7 
Murine 6A6 Fab in complex with humanized anti-Tissue Factor D3H44 Fab 1pkq 
Myelin Oligodendrocyte Glycoprotein-(8-18C5) Fab-complex 1plg 
EVIDENCE FOR THE EXTENDED HELICAL NATURE OF POLYSACCHARIDE EPITOPES. THE 2.8 ANGSTROMS RESOLUTION STRUCTURE AND THERMODYNAMICS OF LIGAND BINDING OF AN ANTIGEN BINDING FRAGMENT SPECIFIC FOR ALPHA-(2->8)-POLYSIALIC ACID 1pqz 
MURINE CYTOMEGALOVIRUS IMMUNOMODULATORY PROTEIN M144 1psk 
THE CRYSTAL STRUCTURE OF AN FAB FRAGMENT THAT BINDS TO THE MELANOMA-ASSOCIATED GD2 GANGLIOSIDE 1pz5 
Structural basis of peptide-carbohydrate mimicry in an antibody combining site 1q0x 
Anti-morphine Antibody 9B1 Unliganded Form 1q0y 
Anti-Morphine Antibody 9B1 Complexed with Morphine 1q1j 
Crystal Structure Analysis of anti-HIV-1 Fab 447-52D in complex with V3 peptide 1q72 
Anti-Cocaine Antibody M82G2 Complexed with Cocaine 1q9k 
S25-2 Fab Unliganded 1 1q9l 
S25-2 Fab Unliganded 2 1q9o 
S45-18 Fab Unliganded 1q9w 
S45-18 Fab pentasaccharide bisphosphate complex 1qbl 
FAB E8 (FABE8A) X-RAY STRUCTURE AT 2.26 ANGSTROM RESOLUTION 1qbm 
FAB E8B ANTIBODY, X-RAY STRUCTURE AT 2.37 ANGSTROMS RESOLUTION 1qfu 
INFLUENZA VIRUS HEMAGGLUTININ COMPLEXED WITH A NEUTRALIZING ANTIBODY 1qgc 
STRUCTURE OF THE COMPLEX OF AN FAB FRAGMENT OF A NEUTRALIZING ANTIBODY WITH FOOT AND MOUTH DISEASE VIRUS 1qkz 
Fab fragment (MN14C11.6) in complex with a peptide antigen derived from Neisseria meningitidis P1.7 serosubtype antigen and domain II from Streptococcal protein G 1qlr 
CRYSTAL STRUCTURE OF THE FAB FRAGMENT OF A HUMAN MONOCLONAL IgM COLD AGGLUTININ 1qrn 
CRYSTAL STRUCTURE OF HUMAN A6 TCR COMPLEXED WITH HLA-A2 BOUND TO ALTERED HTLV-1 TAX PEPTIDE P6A 1qse 
STRUCTURE OF HUMAN A6-TCR BOUND TO HLA-A2 COMPLEXED WITH ALTERED HTLV-1 TAX PEPTIDE V7R 1qsf 
STRUCTURE OF A6-TCR BOUND TO HLA-A2 COMPLEXED WITH ALTERED HTLV-1 TAX PEPTIDE Y8A 1qyg 
ANTI-COCAINE ANTIBODY M82G2 COMPLEXED WITH BENZOYLECGONINE 1r0a 
Crystal structure of HIV-1 reverse transcriptase covalently tethered to DNA template-primer solved to 2.8 angstroms 1r24 
FAB FROM MURINE IGG3 KAPPA 1r3i 
potassium channel KcsA-Fab complex in Rb+ 1r3j 
potassium channel KcsA-Fab complex in high concentration of Tl+ 1r3k 
potassium channel KcsA-Fab complex in low concentration of Tl+ 1r3l 
potassium channel KcsA-Fab complex in Cs+ 1r70 
Model of human IgA2 determined by solution scattering, curve fitting and homology modelling 1rfd 
ANTI-COCAINE ANTIBODY M82G2 1rhf 
Crystal Structure of human Tyro3-D1D2 1rhh 
Crystal Structure of the Broadly HIV-1 Neutralizing Fab X5 at 1.90 Angstrom Resolution 1rih 
Crystal Structure of Fab 14F7, a unique anti-tumor antibody specific for N-glycolyl GM3 1riu 
Anti-Cocaine Antibody M82G2 Complexed With Norbenzoylecgonine 1riv 
Anti-Cocaine Antibody M82G2 Complexed With meta-Oxybenzoylecgonine 1rjl 
Structure of the complex between OspB-CT and bactericidal Fab-H6831 1rmf 
STRUCTURES OF A MONOCLONAL ANTI-ICAM-1 ANTIBODY R6.5 FRAGMENT AT 2.8 ANGSTROMS RESOLUTION 1rpq 
High Affinity IgE Receptor (alpha chain) Complexed with Tight-Binding E131 'zeta' Peptide from Phage Display 1ru9 
Crystal Structure (A) of u.v.-irradiated cationic cyclization antibody 4C6 Fab at pH 4.6 with a data set collected in-house. 1rua 
Crystal structure (B) of u.v.-irradiated cationic cyclization antibody 4C6 fab at pH 4.6 with a data set collected at SSRL beamline 11-1. 1ruk 
Crystal structure (C) of native cationic cyclization antibody 4C6 fab at pH 4.6 with a data set collected at SSRL beamline 9-1 1rul 
Crystal Structure (D) of u.v.-irradiated cationic cyclization antibody 4C6 Fab at pH 5.6 with a data set collected at SSRL beamline 11-1. 1rum 
Crystal structure (F) of H2O2-soaked cationic cyclization antibody 4C6 fab at pH 8.5 with a data set collected at SSRL beamline 9-1. 1rup 
Crystal structure (G) of native cationic cyclization antibody 4C6 fab at pH 8.5 with a data set collected at APS beamline 19-ID 1ruq 
Crystal Structure (H) of u.v.-irradiated Diels-Alder antibody 13G5 Fab at pH 8.0 with a data set collected in house. 1rur 
Crystal Structure (I) of native Diels-Alder antibody 13G5 Fab at pH 8.0 with a data set collected at SSRL beamline 9-1. 1ry7 
Crystal Structure of the 3 Ig form of FGFR3c in complex with FGF1 1rz7 
CRYSTAL STRUCTURE OF HUMAN ANTI-HIV-1 GP120-REACTIVE ANTIBODY 48D 1rz8 
CRYSTAL STRUCTURE OF HUMAN ANTI-HIV-1 GP120-REACTIVE ANTIBODY 17B 1rzf 
Crystal structure of Human anti-HIV-1 GP120-reactive antibody E51 1rzg 
Crystal structure of Human anti-HIV-1 GP120 reactive antibody 412d 1rzi 
Crystal structure of human anti-HIV-1 gp120-reactive antibody 47e fab 1rzj 
HIV-1 HXBC2 GP120 ENVELOPE GLYCOPROTEIN COMPLEXED WITH CD4 AND INDUCED NEUTRALIZING ANTIBODY 17B 1rzk 
HIV-1 YU2 GP120 ENVELOPE GLYCOPROTEIN COMPLEXED WITH CD4 AND INDUCED NEUTRALIZING ANTIBODY 17B 1s3k 
Crystal Structure of a Humanized Fab (hu3S193) in Complex with the Lewis Y Tetrasaccharide 1s5h 
Potassium Channel Kcsa-Fab Complex T75C mutant in K+ 1s5i 
Fab (LNKB-2) of monoclonal antibody to Human Interleukin-2, crystal structure 1s78 
Insights into ErbB signaling from the structure of the ErbB2-pertuzumab complex 1sbb 
T-CELL RECEPTOR BETA CHAIN COMPLEXED WITH SUPERANTIGEN SEB 1sbs 
CRYSTAL STRUCTURE OF AN ANTI-HCG FAB 1seq 
Fab MNAC13 1sm3 
CRYSTAL STRUCTURE OF THE TUMOR SPECIFIC ANTIBODY SM3 COMPLEX WITH ITS PEPTIDE EPITOPE 1sy6 
Crystal Structure of CD3gammaepsilon Heterodimer in Complex with OKT3 Fab Fragment 1t03 
HIV-1 reverse transcriptase crosslinked to tenofovir terminated template-primer (complex P) 1t04 
Three dimensional structure of a humanized anti-IFN-Gamma Fab in C2 space group 1t2q 
The Crystal Structure of an NNA7 Fab that recognizes an N-type blood group antigen 1t3f 
THREE DIMENSIONAL STRUCTURE OF A HUMANIZED ANTI-IFN-GAMMA FAB (HuZAF) IN P21 21 21 SPACE GROUP 1t4k 
Crystal Structure of Unliganded Aldolase Antibody 93F3 Fab 1t66 
The structure of FAB with intermediate affinity for fluorescein. 1t6w 
RATIONAL DESIGN OF A CALCIUM-BINDING ADHESION PROTEIN NMR, 20 STRUCTURES 1t83 
CRYSTAL STRUCTURE OF A HUMAN TYPE III FC GAMMA RECEPTOR IN COMPLEX WITH AN FC FRAGMENT OF IGG1 (ORTHORHOMBIC) 1t89 
CRYSTAL STRUCTURE OF A HUMAN TYPE III FC GAMMA RECEPTOR IN COMPLEX WITH AN FC FRAGMENT OF IGG1 (HEXAGONAL) 1tcr 
MURINE T-CELL ANTIGEN RECEPTOR 2C CLONE 1tet 
CRYSTAL STRUCTURE OF AN ANTICHOLERA TOXIN PEPTIDE COMPLEX AT 2.3 ANGSTROMS 1tjg 
Crystal Structure of the broadly neutralizing anti-HIV-1 antibody 2F5 in complex with a gp41 7mer epitope 1tjh 
Crystal Structure of the broadly neutralizing anti-HIV-1 antibody 2F5 in complex with a gp41 11mer epitope 1tji 
Crystal Structure of the broadly neutralizing anti-HIV-1 antibody 2F5 in complex with a gp41 17mer epitope 1tpx 
Ovine recombinant PrP(114-234), ARQ variant in complex with the Fab of the VRQ14 antibody 1tqb 
Ovine recombinant PrP(114-234), VRQ variant in complex with the Fab of the VRQ14 antibody 1tqc 
Ovine recombinant PrP(114-234), ARR variant in complex with the VRQ14 Fab fragment (IgG2a) 1tzg 
Crystal structure of HIV-1 neutralizing human Fab 4E10 in complex with a 13-residue peptide containing the 4E10 epitope on gp41 1tzh 
Crystal Structure of the Fab YADS1 Complexed with h-VEGF 1tzi 
Crystal Structure of the Fab YADS2 Complexed with h-VEGF 1u58 
Crystal structure of the murine cytomegalovirus MHC-I homolog m144 1u6a 
Crystal Structure of the Broadly Neutralizing Anti-HIV Fab F105 1u8h 
Crystal structure of the HIV-1 Cross Neutralizing Monoclonal Antibody 2F5 in complex with gp41 Peptide ALDKWAS 1u8i 
Crystal structure of the HIV-1 Cross Neutralizing Monoclonal Antibody 2F5 in complex with gp41 Peptide ELDKWAN 1u8j 
Crystal structure of the HIV-1 Cross Neutralizing Monoclonal Antibody 2F5 in complex with gp41 Peptide ELDKWAG 1u8k 
Crystal structure of the HIV-1 Cross Neutralizing Monoclonal Antibody 2F5 in complex with gp41 Peptide LELDKWASL 1u8l 
Crystal structure of the HIV-1 Cross Neutralizing Monoclonal Antibody 2F5 in complex with gp41 Peptide DLDRWAS 1u8m 
Crystal structure of the HIV-1 Cross Neutralizing Monoclonal Antibody 2F5 in complex with gp41 Peptide ELDKYAS 1u8n 
Crystal structure of the HIV-1 Cross Neutralizing Monoclonal Antibody 2F5 in complex with gp41 Peptide ELDKFAS 1u8o 
Crystal structure of the HIV-1 Cross Neutralizing Monoclonal Antibody 2F5 in complex with gp41 Peptide ELDKHAS 1u8p 
Crystal structure of the HIV-1 Cross Neutralizing Monoclonal Antibody 2F5 in complex with gp41 Peptide ECDKWCS 1u8q 
Crystal structure of the HIV-1 Cross Neutralizing Monoclonal Antibody 2F5 in complex with gp41 Peptide ELEKWAS 1u91 
Crystal structure of the HIV-1 Cross Neutralizing Monoclonal Antibody 2F5 in complex with gp41 Peptide Analog ENDKW-[Dap]-S (cyclic) 1u92 
Crystal structure of the HIV-1 Cross Neutralizing Monoclonal Antibody 2F5 in complex with gp41 Peptide Analog E-[Dap]-DKWQS (cyclic) 1u93 
Crystal structure of the HIV-1 Cross Neutralizing Monoclonal Antibody 2F5 in complex with gp41 Peptide Analog EQDKW-[Dap]-S (cyclic) 1u95 
Crystal structure of the HIV-1 Cross Neutralizing Monoclonal Antibody 2F5 in complex with gp41 Peptide ELDHWAS 1ub5 
Crystal structure of Antibody 19G2 with hapten at 100K 1ub6 
Crystal structure of Antibody 19G2 with sera ligand 1ucb 
STRUCTURE OF UNCOMPLEXED FAB COMPARED TO COMPLEX (1CLY, 1CLZ) 1ufu 
Crystal structure of ligand binding domain of immunoglobulin-like transcript 2 (ILT2; LIR-1) 1ugn 
Crystal structure of LIR1.02, one of the alleles of LIR1 1uj3 
Crystal structure of a humanized Fab fragment of anti-tissue-factor antibody in complex with tissue factor 1um4 
Catalytic Antibody 21H3 with hapten 1um5 
Catalytic Antibody 21H3 with alcohol substrate 1um6 
catalytic antibody 21h3 1uwe 
14d9 catalytic antibody 1uwg 
Molecular Mechanism of Enantioselective Proton Transfer to Carbon in Catalytic Antibody 14D9 1uwx 
P1.2 serosubtype antigen derived from N. meningitidis PorA in complex with Fab fragment 1uyw 
Crystal Structure of the antiflavivirus Fab4g2 1uz6 
anti-Lewis X Fab fragment uncomplexed 1uz8 
anti-Lewis X Fab fragment in complex with Lewis X 1v7m 
Human Thrombopoietin Functional Domain Complexed To Neutralizing Antibody TN1 Fab 1v7n 
Human Thrombopoietin Functional Domain Complexed To Neutralizing Antibody TN1 Fab 1vdg 
Crystal structure of LIR1.01, one of the alleles of LIR1 1vge 
TR1.9 FAB FRAGMENT OF A HUMAN IGG1 KAPPA AUTOANTIBODY 1vpo 
Crystal Structure Analysis of the Anti-testosterone Fab in Complex with Testosterone 1w72 
Crystal structure of HLA-A1:MAGE-A1 in complex with Fab-Hyb3 1wc7 
FAB FRAGMENT OF PLP-DEPENDENT CATALYTIC ANTIBODY 15A9 IN COMPLEX WITH PHOSPHOPYRIDOXYL-L-ALANINE 1wcb 
PLP-DEPENDENT CATALYTIC ANTIBODY 15A9 IN COMPLEX WITH ITS HAPTEN 1wej 
IGG1 FAB FRAGMENT (OF E8 ANTIBODY) COMPLEXED WITH HORSE CYTOCHROME C AT 1.8 A RESOLUTION 1wio 
STRUCTURE OF T-CELL SURFACE GLYCOPROTEIN CD4, TETRAGONAL CRYSTAL FORM 1wip 
STRUCTURE OF T-CELL SURFACE GLYCOPROTEIN CD4, MONOCLINIC CRYSTAL FORM 1wiq 
STRUCTURE OF T-CELL SURFACE GLYCOPROTEIN CD4, TRIGONAL CRYSTAL FORM 1xcq 
Complex HCV core-Fab 19D9D6-Protein L mutant (D55A,L57H,Y64W) in space group P21 1xct 
Complex HCV core-Fab 19D9D6-Protein L mutant (D55A, L57H, Y64W) in space group P21212 1xf2 
Structure of Fab DNA-1 complexed with dT3 1xf3 
Structure of ligand-free Fab DNA-1 in space group P65 1xf4 
Structure of ligand-free Fab DNA-1 in space group P321 solved from crystals with perfect hemihedral twinning 1xf5 
Complex HCV core-Fab 19D9D6-Protein L mutant (H74C, Y64W)in space group P21212 1xgp 
Structure for antibody HyHEL-63 Y33A mutant complexed with hen egg lysozyme 1xgq 
Structure for antibody HyHEL-63 Y33V mutant complexed with hen egg lysozyme 1xgr 
Structure for antibody HyHEL-63 Y33I mutant complexed with hen egg lysozyme 1xgt 
Structure for antibody HyHEL-63 Y33L mutant complexed with hen egg lysozyme 1xgu 
Structure for antibody HyHEL-63 Y33F mutant complexed with hen egg lysozyme 1xgy 
Crystal Structure of Anti-Meta I Rhodopsin Fab Fragment K42-41L 1y0l 
Catalytic elimination antibody 34E4 in complex with hapten 1y18 
Fab fragment of catalytic elimination antibody 34E4 E(H50)D mutant in complex with hapten 1ya5 
Crystal structure of the titin domains z1z2 in complex with telethonin 1yec 
STRUCTURE OF A CATALYTIC ANTIBODY IGG2A FAB FRAGMENT (D2.3) 1yed 
STRUCTURE OF A CATALYTIC ANTIBODY IGG2A FAB FRAGMENT (D2.4) 1yee 
STRUCTURE OF A CATALYTIC ANTIBODY, IGG2A FAB FRAGMENT (D2.5) 1yef 
STRUCTURE OF IGG2A FAB FRAGMENT (D2.3) COMPLEXED WITH SUBSTRATE ANALOGUE 1yeg 
STRUCTURE OF IGG2A FAB FRAGMENT (D2.3) COMPLEXED WITH REACTION PRODUCT 1yeh 
STRUCTURE OF IGG2A FAB FRAGMENT 1yei 
CATALYTIC ANTIBODY D2.3 COMPLEX 1yej 
CATALYTIC ANTIBODY COMPLEX 1yek 
CATALYTIC ANTIBODY D2.3 COMPLEX 1yjd 
Crystal structure of human CD28 in complex with the Fab fragment of a mitogenic antibody (5.11A1) 1ymh 
anti-HCV Fab 19D9D6 complexed with protein L (PpL) mutant A66W 1ymm 
TCR/HLA-DR2b/MBP-peptide complex 1ynk 
Identification of Key residues of the NC6.8 Fab antibody fragment binding to synthetic sweeteners: Crystal structure of NC6.8 co-crystalized with high potency sweetener compound SC45647 1ynl 
Identification of Key residues of the NC6.8 Fab antibody fragment binding to synthetic sweeterners: Crystal structure of NC6.8 co-crystalized with high potency sweetener compound SC45647 1ynt 
Structure of the immunodominant epitope displayed by the surface antigen 1 (SAG1) of Toxoplasma gondii complexed to a monoclonal antibody 1ypz 
Immune receptor 1yqv 
The crystal structure of the antibody Fab HyHEL5 complex with lysozyme at 1.7A resolution 1yuh 
FAB FRAGMENT 1yy8 
Crystal structure of the Fab fragment from the monoclonal antibody cetuximab/Erbitux/IMC-C225 1yy9 
Structure of the extracellular domain of the epidermal growth factor receptor in complex with the Fab fragment of cetuximab/Erbitux/IMC-C225 1yyl 
crystal structure of CD4M33, a scorpion-toxin mimic of CD4, in complex with HIV-1 YU2 gp120 envelope glycoprotein and anti-HIV-1 antibody 17b 1yym 
crystal structure of F23, a scorpion-toxin mimic of CD4, in complex with HIV-1 YU2 gp120 envelope glycoprotein and anti-HIV-1 antibody 17b 1z3g 
Crystal structure of complex between Pvs25 and Fab fragment of malaria transmission blocking antibody 2A8 1z7z 
Cryo-em structure of human coxsackievirus A21 complexed with five domain icam-1kilifi 1za3 
The crystal structure of the YSd1 Fab bound to DR5 1za6 
The structure of an antitumor CH2-domain-deleted humanized antibody 1zan 
Crystal structure of anti-NGF AD11 Fab 1zea 
Structure of the anti-cholera toxin antibody Fab fragment TE33 in complex with a D-peptide 1zgl 
Crystal structure of 3A6 TCR bound to MBP/HLA-DR2a 1zls 
FAB 2G12 + Man4 1zlu 
FAB 2G12 + Man5 1zlv 
Fab 2G12 + Man7 1zlw 
Fab 2G12 + Man8 1ztx 
West Nile Virus Envelope Protein DIII in complex with neutralizing E16 antibody Fab 1zvo 
Semi-extended solution structure of human myeloma immunoglobulin D determined by constrained X-ray scattering 1zwi 
Structure of mutant KcsA potassium channel 25c8 
CATALYTIC ANTIBODY 5C8, FAB-HAPTEN COMPLEX 2a1w 
Anti-cocaine antibody 7.5.21, crystal form I 2a38 
Crystal structure of the N-Terminus of titin 2a6d 
Crystal structure analysis of the anti-arsonate germline antibody 36-65 in complex with a phage display derived dodecapeptide RLLIADPPSPRE 2a6i 
Crystal structure analysis of the anti-arsonate germline antibody 36-65 in complex with a phage display derived dodecapeptide KLASIPTHTSPL 2a6j 
Crystal structure analysis of the anti-arsonate germline antibody 36-65 2a6k 
Crystal Structure Analysis of the germline antibody 36-65 Fab in complex with the dodecapeptide SLGDNLTNHNLR 2a77 
Anti-Cocaine Antibody 7.5.21, Crystal Form II 2aab 
Structural basis of antigen mimicry in a clinically relevant melanoma antigen system 2adf 
Crystal Structure and Paratope Determination of 82D6A3, an Antithrombotic Antibody Directed Against the von Willebrand factor A3-Domain 2adg 
Crystal structure of monoclonal anti-CD4 antibody Q425 2adi 
Crystal structure of monoclonal anti-CD4 antibody Q425 in complex with Barium 2adj 
Crystal structure of monoclonal anti-CD4 antibody Q425 in complex with Calcium 2aep 
An epidemiologically significant epitope of a 1998 influenza virus neuraminidase forms a highly hydrated interface in the NA-antibody complex. 2aeq 
An epidemiologically significant epitope of a 1998 influenza virus neuraminidase forms a highly hydrated interface in the NA-antibody complex. 2agj 
Crystal Structure of a glycosylated Fab from an IgM cryoglobulin with properties of a natural proteolytic antibody 2ai0 
Anti-Cocaine Antibody 7.5.21, Crystal Form III 2aj3 
Crystal Structure of a Cross-Reactive HIV-1 Neutralizing CD4-Binding Site Antibody Fab m18 2ajs 
Crystal structure of cocaine catalytic antibody 7A1 Fab' in complex with heptaethylene glycol 2aju 
Cyrstal structure of cocaine catalytic antibody 7A1 Fab' 2ajv 
Crystal Structure of Cocaine catalytic Antibody 7A1 Fab' in Complex with Cocaine 2ajx 
Crystal Structure of Cocaine catalytic Antibody 7A1 Fab' in Complex with Transition State Analog 2ajy 
Crystal Structure of Cocaine catalytic Antibody 7A1 Fab' in Complex with ecgonine methyl ester and benzoic acid 2ajz 
Crystal Structure of Cocaine catalytic Antibody 7A1 Fab' in Complex with ecgonine methyl ester 2ak1 
Crystal Structure of Cocaine catalytic Antibody 7A1 Fab' in Complex with benzoic acid 2ak4 
Crystal Structure of SB27 TCR in complex with HLA-B*3508-13mer peptide 2arj 
CD8alpha-alpha in complex with YTS 105.18 Fab 2atk 
Structure of a mutant KcsA K+ channel 2axh 
Crystal structures of T cell receptor beta chains related to rheumatoid arthritis 2axj 
Crystal structures of T cell receptor beta chains related to rheumatoid arthritis 2b0s 
Crystal structure analysis of anti-HIV-1 V3 Fab 2219 in complex with MN peptide 2b1a 
Crystal structure analysis of anti-HIV-1 V3 Fab 2219 in complex with UG1033 peptide 2b1h 
Crystal structure analysis of anti-HIV-1 V3 Fab 2219 in complex with UG29 peptide 2b2x 
VLA1 RdeltaH I-domain complexed with a quadruple mutant of the AQC2 Fab 2b4c 
Crystal structure of HIV-1 JR-FL gp120 core protein containing the third variable region (V3) complexed with CD4 and the X5 antibody 2bdn 
Crystal structure of human MCP-1 bound to a blocking antibody, 11K2 2bmk 
Fab fragment of PLP-dependent catalytic antibody 15A9 in complex with phosphopyridoxyl-D-alanine 2bnq 
Structural and kinetic basis for heightened immunogenicity of T cell vaccines 2bnr 
Structural and kinetic basis for heightened immunogenicity of T cell vaccines 2bnu 
Structural and kinetic basis for heightened immunogenicity of T cell vaccines 2bob 
Potassium channel KcsA-Fab complex in thallium with tetrabutylammonium (TBA) 2boc 
Potassium channel KcsA-Fab complex in thallium with tetraethylarsonium (TEAs) 2brr 
Complex of the neisserial PorA P1.4 epitope peptide and two Fab- fragments (antibody MN20B9.34) 2c1o 
ENAIIHis Fab fragment in the free form 2c1p 
Fab-fragment of enantioselective antibody complexed with finrozole 2cde 
Structure and binding kinetics of three different human CD1d-alpha- Galactosylceramide specific T cell receptors - iNKT-TCR 2cdf 
Structure and binding kinetics of three different human CD1d-alpha- Galactosylceramide-specific T cell receptors (TCR 5E) 2cdg 
Structure and binding kinetics of three different human CD1d-alpha- Galactosylceramide-specific T cell receptors (TCR 5B) 2cgr 
LOCAL AND TRANSMITTED CONFORMATIONAL CHANGES ON COMPLEXATION OF AN ANTI-SWEETENER FAB 2ch8 
Structure of the Epstein-Barr Virus Oncogene BARF1 2ck0 
ANTI-ANTI-IDIOTYPIC ANTIBODY AGAINST HUMAN ANGIOTENSIN II, COMPLEX WITH A SYNTHETIC CYCLIC PEPTIDE 2ckb 
STRUCTURE OF THE 2C/KB/DEV8 COMPLEX 2cmr 
Crystal structure of the HIV-1 neutralizing antibody D5 Fab bound to the gp41 inner-core mimetic 5-helix 2d03 
Crystal structure of the G91S mutant of the NNA7 Fab 2d3v 
Crystal Structure of Leukocyte Ig-like Receptor A5 (LILRA5/LIR9/ILT11) 2dbl 
MOLECULAR BASIS OF CROSS-REACTIVITY AND THE LIMITS OF ANTIBODY-ANTIGEN COMPLEMENTARITY 2dd8 
Crystal Structure of SARS-CoV Spike Receptor-Binding Domain Complexed with Neutralizing Antibody 2ddq 
Crystal Structure of the Fab fragment of a R310 antibody complexed with (R)-HNE-histidine adduct 2dl2 
KILLER IMMUNOGLOBULIN RECEPTOR 2DL2 2dli 
KILLER IMMUNOGLOBULIN RECEPTOR 2DL2,TRIGONAL FORM 2dqt 
High resolution crystal structure of the complex of the hydrolytic antibody Fab 6D9 and a transition-state analog 2dqu 
Crystal form II: high resolution crystal structure of the complex of the hydrolytic antibody Fab 6D9 and a transition-state analog 2dtm 
Thermodynamic and structural analyses of hydrolytic mechanism by catalytic antibodies 2dts 
Crystal Structure of the Defucosylated Fc Fragment from Human Immunoglobulin G1 2dwd 
crystal structure of KcsA-FAB-TBA complex in Tl+ 2dwe 
Crystal structure of KcsA-FAB-TBA complex in Rb+ 2e9w 
Crystal structure of the extracellular domain of Kit in complex with stem cell factor (SCF) 2ec8 
Crystal structure of the exctracellular domain of the receptor tyrosine kinase, Kit 2eh7 
Crystal structure of humanized KR127 FAB 2eh8 
Crystal structure of the complex of humanized KR127 fab and PRES1 peptide epitope 2esg 
Solution structure of the complex between immunoglobulin IgA1 and human serum albumin 2esv 
Structure of the HLA-E-VMAPRTLIL/KK50.4 TCR complex 2exw 
Crystal structure of a EcClC-Fab complex in the absence of bound ions 2exy 
Crystal structure of the E148Q Mutant of EcClC, Fab complexed in absence of bound ions 2eyr 
A structural basis for selection and cross-species reactivity of the semi-invariant NKT cell receptor in CD1d/glycolipid recognition 2eys 
A structural basis for selection and cross-species reactivity of the semi-invariant NKT cell receptor in CD1d/glycolipid recognition 2eyt 
A structural basis for selection and cross-species reactivity of the semi-invariant NKT cell receptor in CD1d/glycolipid recognition 2ez0 
Crystal structure of the S107A/E148Q/Y445A mutant of EcClC, in complex with a FaB fragment 2f19 
THREE-DIMENSIONAL STRUCTURE OF TWO CRYSTAL FORMS OF FAB R19.9, FROM A MONOCLONAL ANTI-ARSONATE ANTIBODY 2f53 
Directed Evolution of Human T-cell Receptor CDR2 residues by phage display dramatically enhances affinity for cognate peptide-MHC without apparent cross-reactivity 2f54 
Directed evolution of human T cell receptor CDR2 residues by phage display dramatically enhances affinity for cognate peptide-MHC without increasing apparent cross-reactivity 2f58 
IGG1 FAB FRAGMENT (58.2) COMPLEX WITH 12-RESIDUE CYCLIC PEPTIDE (INCLUDING RESIDUES 315-324 OF HIV-1 GP120) (MN ISOLATE) 2f5a 
CRYSTAL STRUCTURE OF FAB' FROM THE HIV-1 NEUTRALIZING ANTIBODY 2F5 2f5b 
CRYSTAL STRUCTURE OF FAB' FROM THE HIV-1 NEUTRALIZING ANTIBODY 2F5 IN COMPLEX WITH ITS GP41 EPITOPE 2f8v 
Structure of full length telethonin in complex with the N-terminus of titin 2fat 
An anti-urokinase plasminogen activator receptor (UPAR) antibody: Crystal structure and binding epitope 2fb4 
DIR PRIMAERSTRUKTUR DES KRISTALLISIERBAREN MONOKLONALEN IMMUNOGLOBULINS IGG1 KOL. II. AMINOSAEURESEQUENZ DER L-KETTE, LAMBDA-TYP, SUBGRUPPE I (GERMAN) 2fbj 
REFINED CRYSTAL STRUCTURE OF THE GALACTAN-BINDING IMMUNOGLOBULIN FAB J539 AT 1.95-ANGSTROMS RESOLUTION 2fd6 
Structure of Human Urokinase Plasminogen Activator in Complex with Urokinase Receptor and an anti-upar antibody at 1.9 A 2fec 
Structure of the E203Q mutant of the Cl-/H+ exchanger CLC-ec1 from E.Coli 2fed 
Structure of the E203Q mutant of the Cl-/H+ exchanger CLC-ec1 from E.Coli 2fee 
Structure of the Cl-/H+ exchanger CLC-ec1 from E.Coli in NaBr 2fgw 
X-RAY STRUCTURES OF FRAGMENTS FROM BINDING AND NONBINDING VERSIONS OF A HUMANIZED ANTI-CD18 ANTIBODY: STRUCTURAL INDICATIONS OF THE KEY ROLE OF VH RESIDUES 59 TO 65 2fjf 
Structure of the G6 Fab, a phage derived VEGF binding Fab 2fjg 
Structure of the G6 Fab, a phage derived Fab fragment, in complex with VEGF 2fjh 
Structure of the B20-4 Fab, a phage derived Fab fragment, in complex with VEGF 2fl5 
Cofactor-containing antibodies: Crystal structure of the original yellow antibody 2fr4 
Structure of Fab DNA-1 complexed with a stem-loop DNA ligand 2fx7 
Crystal structure of hiv-1 neutralizing human fab 4e10 in complex with a 16-residue peptide encompassing the 4e10 epitope on gp41 2fx8 
Crystal structure of hiv-1 neutralizing human fab 4e10 in complex with an aib-induced peptide encompassing the 4e10 epitope on gp41 2fx9 
Crystal structure of hiv-1 neutralizing human fab 4e10 in complex with a thioether-linked peptide encompassing the 4e10 epitope on gp41 2g2r 
Green-fluorescent antibody 11G10 in complex with its hapten (nitro-stilbene derivative) 2g5b 
Crystal Structure of the anti-Bax monoclonal antibody 6A7 and a Bax peptide. 2g60 
Structure of anti-FLAG M2 Fab domain 2g75 
Crystal Structure of anti-SARS m396 Antibody 2gcy 
humanized antibody C25 Fab fragment 2gfb 
CRYSTAL STRUCTURE OF A CATALYTIC FAB HAVING ESTERASE-LIKE ACTIVITY 2gj6 
The complex between TCR A6 and human Class I MHC HLA-A2 with the modified HTLV-1 TAX (Y5K-4-[3-Indolyl]-butyric acid) peptide 2gj7 
Crystal Structure of a gE-gI/Fc complex 2gjz 
Structure of Catalytic Elimination Antibody 13G5 from a crystal in space group P2(1) 2gk0 
Structure of Catalytic Elimination Antibody 13G5 from a twinned crystal in space group C2 2gsi 
Crystal Structure of a Murine Fab in Complex with an 11 Residue Peptide Derived from Staphylococcal Nuclease 2gy5 
Tie2 Ligand-Binding Domain Crystal Structure 2gy7 
Angiopoietin-2/Tie2 Complex Crystal Structure 2h1p 
THE THREE-DIMENSIONAL STRUCTURES OF A POLYSACCHARIDE BINDING ANTIBODY TO CRYPTOCOCCUS NEOFORMANS AND ITS COMPLEX WITH A PEPTIDE FROM A PHAGE DISPLAY LIBRARY: IMPLICATIONS FOR THE IDENTIFICATION OF PEPTIDE MIMOTOPES 2h2p 
Crystal structure of CLC-ec1 in complex with Fab fragment in SeCN- 2h2s 
Crystal Structure of E148A mutant of CLC-ec1 in SeCN- 2h32 
Crystal structure of the pre-B cell receptor 2h8p 
Structure of a K channel with an amide to ester substitution in the selectivity filter 2h9g 
Crystal structure of phage derived Fab BdF1 with human Death Receptor 5 (DR5) 2hfe 
Rb+ complex of a K channel with an amide to ester substitution in the selectivity filter 2hff 
Crystal structure of CB2 Fab 2hfg 
Crystal structure of hBR3 bound to CB3s-Fab 2hg5 
Cs+ complex of a K channel with an amide to ester substitution in the selectivity filter 2hh0 
Structure of an Anti-PrP Fab, P-Clone, in Complex with its Cognate Bovine Peptide Epitope. 2hjf 
Potassium channel kcsa-fab complex with tetrabutylammonium (TBA) 2hkf 
Crystal structure of the Complex Fab M75- Peptide 2hkh 
Crystal structure of the Fab M75 2hlf 
Structure of the Escherichis coli ClC chloride channel Y445E mutant and Fab complex 2hmi 
HIV-1 REVERSE TRANSCRIPTASE/FRAGMENT OF FAB 28/DNA COMPLEX 2hrp 
ANTIGEN-ANTIBODY COMPLEX 2ht2 
Structure of the Escherichia coli ClC chloride channel Y445H mutant and Fab complex 2ht3 
Structure of the Escherichia coli ClC chloride channel Y445L mutant and Fab complex 2ht4 
Structure of the Escherichia coli ClC chloride channel Y445W mutant and Fab complex 2htk 
Structure of the Escherichia coli ClC chloride channel Y445A mutant and Fab complex 2htl 
Structure of the Escherichia coli ClC chloride channel Y445F mutant and Fab complex 2hvj 
Crystal structure of KcsA-Fab-TBA complex in low K+ 2hvk 
crystal structure of the KcsA-Fab-TBA complex in high K+ 2hwz 
Fab fragment of Humanized anti-viral antibody MEDI-493 (Synagis TM) 2i5y 
Crystal structure of CD4M47, a scorpion-toxin mimic of CD4, in complex with HIV-1 YU2 GP120 envelope glycoprotein and anti-HIV-1 antibody 17B 2i60 
Crystal structure of [Phe23]M47, a scorpion-toxin mimic of CD4, in complex with HIV-1 YU2 GP120 envelope glycoprotein and anti-HIV-1 antibody 17B 2i9l 
Structure of Fab 7D11 from a neutralizing antibody against the poxvirus L1 protein 2ial 
Structural basis for recognition of mutant self by a tumor-specific, MHC class II-restricted TCR 2iam 
Structural basis for recognition of mutant self by a tumor-specific, MHC class II-restricted TCR 2ian 
Structural basis for recognition of mutant self by a tumor-specific, MHC class II-restricted TCR 2iff 
STRUCTURE OF AN ANTIBODY-LYSOZYME COMPLEX: EFFECT OF A CONSERVATIVE MUTATION 2ig2 
DIR PRIMAERSTRUKTUR DES KRISTALLISIERBAREN MONOKLONALEN IMMUNOGLOBULINS IGG1 KOL. II. AMINOSAEURESEQUENZ DER L-KETTE, LAMBDA-TYP, SUBGRUPPE I (GERMAN) 2igf 
CRYSTAL STRUCTURES OF AN ANTIBODY TO A PEPTIDE AND ITS COMPLEX WITH PEPTIDE ANTIGEN AT 2.8 ANGSTROMS 2ih1 
Ion selectivity in a semi-synthetic K+ channel locked in the conductive conformation 2ih3 
Ion selectivity in a semi-synthetic K+ channel locked in the conductive conformation 2ipt 
PFA1 Fab Fragment 2ipu 
PFA1 Fab fragment complexed with Abeta 1-8 peptide 2iq9 
PFA2 FAB fragment, triclinic apo form 2iqa 
PFA2 FAB fragment, monoclinic apo form 2itc 
Potassium Channel KcsA-Fab complex in Sodium Chloride 2itd 
Potassium Channel KcsA-Fab complex in Barium Chloride 2iwg 
COMPLEX BETWEEN THE PRYSPRY DOMAIN OF TRIM21 AND IGG FC 2j4w 
Structure of a Plasmodium vivax apical membrane antigen 1-Fab F8.12. 18 complex 2j5l 
Structure of a Plasmodium falciparum apical membrane antigen 1-Fab F8. 12.19 complex 2j6e 
Crystal Structure of an Autoimmune Complex between a Human IgM Rheumatoid Factor and IgG1 Fc reveals a Novel Fc Epitope and Evidence for Affinity Maturation 2j88 
Hyaluronidase in complex with a monoclonal IgG Fab fragment 2j8u 
Large CDR3a loop alteration as a function of MHC mutation. 2jb5 
Fab fragment in complex with small molecule hapten, crystal form-1 2jb6 
Fab fragment in complex with small molecule hapten, crystal form-2 2jcc 
AH3 recognition of mutant HLA-A2 W167A 2jel 
JEL42 FAB/HPR COMPLEX 2jix 
Crystal structure of ABT-007 FAB fragment with the soluble domain of EPO receptor 2jjs 
Structure of human cd47 in complex with human signal regulatory protein (SIRP) alpha 2jjt 
Structure of human cd47 in complex with human signal regulatory protein (SIRP) alpha 2jk5 
Potassium Channel KcsA in complex with Tetrabutylammonium in high K 2ltq 
High resolution structure of DsbB C41S by joint calculation with solid-state NMR and X-ray data 2mcg 
THREE-DIMENSIONAL STRUCTURE OF A LIGHT CHAIN DIMER CRYSTALLIZED IN WATER. CONFORMATIONAL FLEXIBILITY OF A MOLECULE IN TWO CRYSTAL FORMS 2mcp 
REFINED CRYSTAL STRUCTURE OF THE MC/PC603 FAB-PHOSPHOCHOLINE COMPLEX AT 3.1 ANGSTROMS RESOLUTION 2mim 
NMR structure of the chicken CD3 epsilon delta/gamma heterodimer 2mkl 
2MKL 2mpa 
BACTERICIDAL ANTIBODY AGAINST NEISSERIA MENINGITIDIS 2nlj 
Potassium Channel KcsA(M96V)-Fab complex in KCl 2nr6 
Crystal structure of the complex of antibody and the allergen Bla g 2 2ntf 
Crystal Structure of a Quorum-Quenching Antibody in Complex with an N-Acyl-L-Homoserine Lactone Analog 2nts 
Crystal Structure of SEK-hVb5.1 2nw2 
Crystal structure of ELS4 TCR at 1.4A 2nx5 
Crystal structure of ELS4 TCR bound to HLA-B*3501 presenting EBV peptide EPLPQGQLTAY at 1.7A 2nxy 
HIV-1 gp120 Envelope Glycoprotein(S334A) Complexed with CD4 and Antibody 17b 2nxz 
HIV-1 gp120 Envelope Glycoprotein (T257S, S334A, S375W) Complexed with CD4 and Antibody 17b 2ny0 
HIV-1 gp120 Envelope Glycoprotein (M95W, W96C, T257S, V275C, S334A, S375W, A433M) Complexed with CD4 and Antibody 17b 2ny1 
HIV-1 gp120 Envelope Glycoprotein (I109C, T257S, S334A, S375W, Q428C) Complexed with CD4 and Antibody 17b 2ny2 
HIV-1 gp120 Envelope Glycoprotein (T123C, T257S, S334A, S375W, G431C) Complexed with CD4 and Antibody 17b 2ny3 
HIV-1 gp120 Envelope Glycoprotein (K231C, T257S, E267C, S334A, S375W) Complexed with CD4 and Antibody 17b 2ny4 
HIV-1 gp120 Envelope Glycoprotein (K231C, T257S, E268C, S334A, S375W) Complexed with CD4 and Antibody 17b 2ny5 
HIV-1 gp120 Envelope Glycoprotein (M95W, W96C, I109C, T257S, V275C, S334A, S375W, Q428C, A433M) Complexed with CD4 and Antibody 17b 2ny6 
HIV-1 gp120 Envelope Glycoprotein (M95W, W96C, I109C, T123C, T257S, V275C,S334A, S375W, Q428C, G431C) Complexed with CD4 and Antibody 17b 2ny7 
HIV-1 gp120 Envelope Glycoprotein Complexed with the Broadly Neutralizing CD4-Binding-Site Antibody b12 2nyy 
Crystal structure of botulinum neurotoxin type A complexed with monoclonal antibody CR1 2nz9 
Crystal structure of botulinum neurotoxin type A complexed with monoclonal antibody AR2 2o26 
Structure of a class III RTK signaling assembly 2o5x 
Crystal structure of 1E9 LeuH47Trp/ArgH100Trp, an engineered Diels-Alderase Fab with nM steroid-binding affinity 2o5y 
Crystal structure of the 1E9 LeuH47Trp/ArgH100Trp Fab progesterone complex 2o5z 
Crystal structure of the 1E9 LeuH47Trp/ArgH100Trp Fab 5-beta-androstane-3,17-dione complex 2ojz 
Anti-DNA antibody ED10 2ok0 
Fab ED10-DNA complex 2old 
Bence Jones KWR Protein- Immunoglobulin Light Chain Dimer, P3(2)21 Crystal Form 2om5 
N-Terminal Fragment of Human TAX1 2omb 
Bence Jones KWR Protein- Immunoglobulin Light Chain Dimer, P3(1)21 Crystal Form 2omn 
Bence Jones KWR Protein- Immunoglobulin Light Chain Dimer, P4(3)2(1)2 Crystal Form 2op4 
Crystal Structure of Quorum-Quenching Antibody 1G9 2oqj 
Crystal structure analysis of Fab 2G12 in complex with peptide 2G12.1 2or9 
The structure of the anti-c-myc antibody 9E10 Fab fragment/epitope peptide complex reveals a novel binding mode dominated by the heavy chain hypervariable loops 2orb 
The structure of the anti-c-myc antibody 9E10 Fab fragment 2osl 
Crystal structure of Rituximab Fab in complex with an epitope peptide 2otp 
Crystal Structure of Immunoglobulin-Like Transcript 1 (ILT1/LIR7/LILRA2) 2oz4 
Structural Plasticity in IgSF Domain 4 of ICAM-1 Mediates Cell Surface Dimerization 2p5e 
Crystal Structures of High Affinity Human T-Cell Receptors Bound to pMHC Reveal Native Diagonal Binding Geometry 2p5w 
Crystal structures of high affinity human T-cell receptors bound to pMHC reveal native diagonal binding geometry 2p7t 
Crystal Structure of KcsA mutant 2p8l 
Crystal structure of the HIV-1 Cross Neutralizing Monoclonal Antibody 2F5 in complex with gp41 Peptide ELLELDKWASLWN 2p8m 
Crystal structure of the HIV-1 Cross Neutralizing Monoclonal Antibody 2F5 in complex with gp41 Peptide ELLELDKWASLWN in new crystal form 2p8p 
Crystal structure of the HIV-1 Cross Neutralizing Monoclonal Antibody 2F5 in complex with gp41 Peptide LELDKWASLW[N-Ac] 2pcp 
ANTIBODY FAB COMPLEXED WITH PHENCYCLIDINE 2pet 
Lutheran glycoprotein, N-terminal domains 1 and 2. 2pf6 
Lutheran glycoprotein, N-terminal domains 1 and 2 2po6 
Crystal structure of CD1d-lipid-antigen complexed with Beta-2-Microglobulin, NKT15 Alpha-Chain and NKT15 Beta-Chain 2pr4 
Crystal Structure of Fab' from the HIV-1 Neutralizing Antibody 2F5 2pw1 
Crystal structure of the HIV-1 Cross Neutralizing Monoclonal Antibody 2F5 in complex with gp41 Peptide ELDKWNSL 2pw2 
Crystal structure of the HIV-1 Cross Neutralizing Monoclonal Antibody 2F5 in complex with gp41 Peptide ELDKWKSL 2pye 
Crystal Structures of High Affinity Human T-Cell Receptors Bound to pMHC RevealNative Diagonal Binding Geometry TCR Clone C5C1 Complexed with MHC 2pyf 
Crystal Structures of High Affinity Human T-Cell Receptors Bound to pMHC RevealNative Diagonal Binding Geometry Unbound TCR Clone 5-1 2q76 
Mouse anti-hen egg white lysozyme antibody F10.6.6 Fab fragment 2q86 
Structure of the mouse invariant NKT cell receptor Valpha14 2q8a 
Structure of the malaria antigen AMA1 in complex with a growth-inhibitory antibody 2q8b 
Structure of the malaria antigen AMA1 in complex with a growth-inhibitory antibody 2qad 
Structure of tyrosine-sulfated 412d antibody complexed with HIV-1 YU2 gp120 and CD4 2qhr 
Crystal structure of the 13F6-1-2 Fab fragment bound to its Ebola virus glycoprotein peptide epitope. 2ql1 
Structural Characterization of a Mutated, ADCC-Enhanced Human Fc Fragment 2qqk 
Neuropilin-2 a1a2b1b2 Domains in Complex with a Semaphorin-Blocking Fab 2qql 
Neuropilin-2 a1a2b1b2 Domains in Complex with a Semaphorin-Blocking Fab 2qqn 
Neuropilin-1 b1 Domain in Complex with a VEGF-Blocking Fab 2qr0 
Structure of VEGF complexed to a Fab containing TYR and SER in the CDRs 2qsc 
Crystal structure analysis of anti-HIV-1 V3-Fab F425-B4e8 in complex with a V3-peptide 2qtj 
Solution structure of human dimeric immunoglobulin A 2r0k 
Protease domain of HGFA with inhibitor Fab58 2r0l 
Short Form HGFA with Inhibitory Fab75 2r0w 
PFA2 FAB complexed with Abeta1-8 2r0z 
PFA1 FAB complexed with GripI peptide fragment 2r1w 
Crystal structure of S25-2 Fab in complex with Kdo analogues 2r1x 
Crystal structure of S25-2 Fab in complex with Kdo analogues 2r1y 
Crystal structure of S25-2 Fab in complex with Kdo analogues 2r23 
Crystal structure of S25-2 Fab in complex with Kdo analogues 2r29 
Neutralization of dengue virus by a serotype cross-reactive antibody elucidated by cryoelectron microscopy and x-ray crystallography 2r2b 
Crystal structure of S25-2 Fab in complex with Kdo analogues 2r2e 
Crystal structure of S25-2 Fab in complex with Kdo analogues 2r2h 
Structure of S25-2 in Complex with Ko 2r4r 
Crystal structure of the human beta2 adrenoceptor 2r4s 
Crystal structure of the human beta2 adrenoceptor 2r56 
Crystal Structure of a Recombinant IgE Fab Fragment in Complex with Bovine Beta-Lactoglobulin Allergen 2r69 
Crystal structure of Fab 1A1D-2 complexed with E-DIII of Dengue virus at 3.8 angstrom resolution 2r6p 
Fit of E protein and Fab 1A1D-2 into 24 angstrom resolution cryoEM map of Fab complexed with dengue 2 virus. 2r8s 
High resolution structure of a specific synthetic FAB bound to P4-P6 RNA ribozyme domain 2r9h 
Crystal Structure of Q207C Mutant of CLC-ec1 in complex with Fab 2rcj 
Solution structure of human Immunoglobulin M 2rcs 
IMMUNOGLOBULIN 48G7 GERMLINE FAB-AFFINITY MATURATION OF AN ESTEROLYTIC ANTIBODY 2rgs 
FC-fragment of monoclonal antibody IGG2B from Mus musculus 2uwe 
Large CDR3a loop alteration as a function of MHC mutation 2uyl 
Crystal structure of a monoclonal antibody directed against an antigenic determinant common to Ogawa and Inaba serotypes of Vibrio cholerae O1 2v17 
Structure of the complex of antibody MN423 with a fragment of tau protein 2v5m 
Structural basis for Dscam isoform specificity 2v5r 
Structural basis for Dscam isoform specificity 2v5s 
Structural basis for Dscam isoform specificity 2v7h 
Crystal structure of an immunogen specific anti-mannopyranoside monoclonal antibody Fab fragment 2v7n 
Unusual twinning in crystals of the CitS binding antibody Fab fragment f3p4 2vc2 
Re-refinement of Integrin AlphaIIbBeta3 Headpiece Bound to Antagonist L-739758 2vdk 
Re-refinement of Integrin AlphaIIbBeta3 Headpiece 2vdl 
Re-refinement of Integrin AlphaIIbBeta3 Headpiece 2vdm 
Re-refinement of Integrin AlphaIIbBeta3 Headpiece Bound to Antagonist Tirofiban 2vdn 
Re-refinement of Integrin AlphaIIbBeta3 Headpiece Bound to Antagonist Eptifibatide 2vdo 
Integrin AlphaIIbBeta3 Headpiece Bound to Fibrinogen Gamma chain peptide, HHLGGAKQAGDV 2vdp 
Integrin AlphaIIbBeta3 Headpiece Bound to Fibrinogen Gamma chain peptide,LGGAKQAGDV 2vdq 
Integrin AlphaIIbBeta3 Headpiece Bound to a Chimeric Fibrinogen Gamma chain peptide, HHLGGAKQRGDV 2vdr 
Integrin AlphaIIbBeta3 Headpiece Bound to Fibrinogen Gamma chain chimera peptide, LGGAKQRGDV 2vir 
INFLUENZA VIRUS HEMAGGLUTININ COMPLEXED WITH A NEUTRALIZING ANTIBODY 2vis 
INFLUENZA VIRUS HEMAGGLUTININ, (ESCAPE) MUTANT WITH THR 131 REPLACED BY ILE, COMPLEXED WITH A NEUTRALIZING ANTIBODY 2vit 
INFLUENZA VIRUS HEMAGGLUTININ, MUTANT WITH THR 155 REPLACED BY ILE, COMPLEXED WITH A NEUTRALIZING ANTIBODY 2vl5 
Structure of anti-collagen type II FAb CIIC1 2vlj 
The Structural Dynamics and Energetics of an Immunodominant T-cell Receptor are Programmed by its Vbeta Domain 2vlk 
The Structural Dynamics and Energetics of an Immunodominant T-cell Receptor are Programmed by its Vbeta Domain 2vlm 
The Structural Dynamics and Energetics of an Immunodominant T-cell Receptor are Programmed by its Vbeta Domain 2vlr 
The Structural Dynamics and Energetics of an Immunodominant T-cell Receptor are Programmed by its Vbeta Domain 2vq1 
anti trimeric Lewis X Fab54-5C10-A 2vsc 
Structure of the immunoglobulin-superfamily ectodomain of human CD47 2vwe 
Crystal Structure of Vascular Endothelial Growth Factor-B in Complex with a Neutralizing Antibody Fab Fragment 2vxq 
Crystal structure of the major grass pollen allergen Phl p 2 in complex with its specific IgE-Fab 2vxs 
Structure of IL-17A in complex with a potent, fully human neutralising antibody 2vxt 
Crystal structure of human IL-18 complexed to murine reference antibody 125-2H Fab 2vxu 
Crystal structure of murine reference antibody 125-2H Fab fragment 2vxv 
Crystal structure of human IgG ABT-325 Fab Fragment 2w0f 
Potassium Channel KcsA-Fab Complex with Tetraoctylammonium 2w60 
Anti citrullinated Collagen type 2 antibody acc4 2w65 
Anti citrullinated Collagen type 2 antibody acc4 in complex with a citrullinated peptide 2w9d 
Structure of Fab fragment of the ICSM 18 - anti-Prp therapeutic antibody at 1.65 A resolution. 2w9e 
Structure of ICSM 18 (anti-Prp therapeutic antibody) Fab fragment complexed with human Prp fragment 119-231 2wah 
Crystal Structure of an IgG1 Fc Glycoform (Man9GlcNAc2) 2wbj 
TCR complex 2wng 
complete extracellular structure of human signal regulatory protein ( SIRP) alpha 2wub 
Crystal structure of HGFA in complex with the allosteric non- inhibitory antibody Fab40.deltaTrp 2wuc 
Crystal structure of HGFA in complex with the allosteric non- inhibitory antibody Fab40.deltaTrp and Ac-KQLR-chloromethylketone 2wv3 
Neuroplastin-55 binds to and signals through the fibroblast growth factor receptor 2x1w 
Crystal Structure of VEGF-C in Complex with Domains 2 and 3 of VEGFR2 2x1x 
Crystal Structure of VEGF-C in Complex with Domains 2 and 3 of VEGFR2 in a Tetragonal Crystal Form 2x7l 
Implications of the HIV-1 Rev dimer structure at 3.2A resolution for multimeric binding to the Rev response element 2xa8 
Crystal structure of the Fab domain of omalizumab at 2.41A 2xkn 
Crystal structure of the Fab fragment of the anti-EGFR antibody 7A7 2xn9 
Crystal structure of the ternary complex between human T cell receptor, staphylococcal enterotoxin H and human major histocompatibility complex class II 2xna 
Crystal structure of the complex between human T cell receptor and staphylococcal enterotoxin 2xqb 
Crystal Structure of anti-IL-15 Antibody in Complex with human IL-15 2xqy 
Crystal structure of Pseudorabies core fragment of glycoprotein H in complex with Fab d6.3 2xra 
crystal structure of the HK20 Fab in complex with a gp41 mimetic 5- Helix 2xwt 
Crystal structure of the TSH receptor in complex with a blocking type TSHR autoantibody 2xza 
Crystal Structure of recombinant A.17 antibody FAB fragment 2xzc 
Crystal Structure of phosphonate-modified recombinant A.17 antibody FAB fragment 2xzq 
CRYSTAL STRUCTURE ANALYSIS OF THE ANTI-(4-HYDROXY-3-NITROPHENYL)- ACETYL MURINE GERMLINE MONOCLONAL ANTIBODY BBE6.12H3 FAB FRAGMENT IN COMPLEX WITH A PHAGE DISPLAY DERIVED DODECAPEPTIDE YQLRPNAETLRF 2y06 
CRYSTAL STRUCTURE ANALYSIS OF THE ANTI-(4-HYDROXY-3-NITROPHENYL) - ACETYL MURINE GERMLINE ANTIBODY BBE6.12H3 FAB FRAGMENT IN COMPLEX WITH A PHAGE DISPLAY DERIVED DODECAPEPTIDE GDPRPSYISHLL 2y07 
CRYSTAL STRUCTURE ANALYSIS OF THE ANTI-(4-HYDROXY-3-NITROPHENYL) - ACETYL MURINE GERMLINE MONOCLONAL ANTIBODY BBE6.12H3 FAB FRAGMENT IN COMPLEX WITH A PHAGE DISPLAY DERIVED DODECAPEPTIDE PPYPAWHAPGNI 2y23 
CRYSTAL STRUCTURE OF THE MYOMESIN DOMAINS MY9-MY11 2y25 
Crystal structure of the myomesin domains My11-My13 2y36 
Crystal structure analysis of the anti-(4-hydroxy-3-nitrophenyl)- acetyl murine germline antibody BBE6.12H3 Fab fragment in complex with a phage display derived dodecapeptide DLWTTAIPTIPS 2y5t 
Crystal structure of the pathogenic autoantibody CIIC1 in complex with the triple-helical C1 peptide 2y6s 
Structure of an Ebolavirus-protective antibody in complex with its mucin-domain linear epitope 2y7q 
The high-affinity complex between IgE and its receptor Fc epsilon RI 2yk1 
Structure of human anti-nicotine Fab fragment in complex with nicotine 2ykl 
Structure of human anti-nicotine Fab fragment in complex with nicotine-11-yl-methyl-(4-ethylamino-4-oxo)-butanoate 2ymx 
Crystal structure of inhibitory anti-AChE Fab408 2ypl 
Structural features underlying T-cell receptor sensitivity to concealed MHC class I micropolymorphisms 2ypv 
Crystal structure of the Meningococcal vaccine antigen factor H binding protein in complex with a bactericidal antibody 2z4q 
Crystal structure of a murine antibody FAB 528 2z91 
Crystal structure of the Fab fragment of anti-ciguatoxin antibody 10C9 2z92 
Crystal structure of the Fab fragment of anti-ciguatoxin antibody 10C9 in complex with CTX3C_ABCDE 2z93 
Crystal structure of Fab fragment of anti-ciguatoxin antibody 10C9 in complex with CTX3C-ABCD 2zch 
Crystal structure of human prostate specific antigen complexed with an activating antibody 2zck 
Crystal structure of a ternary complex between PSA, a substrat-acyl intermediate and an activating antibody 2zcl 
Crystal structure of human prostate specific antigen complexed with an activating antibody 2zg1 
Crystal Structure of Two N-terminal Domains of Siglec-5 in Complex with 6'-Sialyllactose 2zg2 
Crystal Structure of Two N-terminal Domains of Native Siglec-5 2zg3 
Crystal Structure of Two N-terminal Domains of Native Siglec-5 in Complex with 3'-Sialyllactose 2zjs 
Crystal Structure of SecYE translocon from Thermus thermophilus with a Fab fragment 2zkh 
Human thrombopoietin neutralizing antibody TN1 FAB 2zpk 
Crystal structure of P20.1 Fab fragment in complex with its antigen peptide 2zuq 
Crystal structure of DsbB-Fab complex 32c2 
STRUCTURE OF AN ACTIVITY SUPPRESSING FAB FRAGMENT TO CYTOCHROME P450 AROMATASE 35c8 
CATALYTIC ANTIBODY 5C8, FAB-INHIBITOR COMPLEX 3aaz 
Crystal structure of the humanized recombinant Fab fragment of a murine; antibody 3agv 
Crystal structure of a human IgG-aptamer complex 3alp 
Cell adhesion protein 3arb 
Ternary crystal structure of the NKT TCR-CD1d-alpha-galactosylceramide analogue-OCH 3ard 
Ternary crystal structure of the mouse NKT TCR-CD1d-3'deoxy-alpha-galactosylceramide 3are 
Ternary crystal structure of the mouse NKT TCR-CD1d-4'deoxy-alpha-galactosylceramide 3arf 
Ternary crystal structure of the mouse NKT TCR-CD1d-C20:2 3arg 
Ternary crystal structure of the mouse NKT TCR-CD1d-alpha-glucosylceramide(C20:2) 3ave 
Crystal Structure of the Fucosylated Fc Fragment from Human Immunoglobulin G1 3axl 
Murine Valpha 10 Vbeta 8.1 T-cell receptor 3ay4 
Crystal structure of nonfucosylated Fc complexed with bis-glycosylated soluble form of Fc gamma receptor IIIa 3b2u 
Crystal structure of isolated domain III of the extracellular region of the epidermal growth factor receptor in complex with the Fab fragment of IMC-11F8 3b2v 
Crystal structure of the extracellular region of the epidermal growth factor receptor in complex with the Fab fragment of IMC-11F8 3b9k 
Crystal structure of CD8alpha-beta in complex with YTS 156.7 FAB 3bae 
Crystal structure of Fab WO2 bound to the N terminal domain of Amyloid beta peptide (1-28) 3bdy 
Dual specific bH1 Fab in complex with VEGF 3be1 
Dual specific bH1 Fab in complex with the extracellular domain of HER2/ErbB-2 3bgf 
X-ray crystal structure of the SARS coronavirus spike receptor binding domain in complex with F26G19 Fab 3bik 
Crystal Structure of the PD-1/PD-L1 Complex 3bis 
Crystal Structure of the PD-L1 3bjl 
LOC, A LAMBDA 1 TYPE LIGHT-CHAIN DIMER (BENCE-JONES PROTEIN) CRYSTALLIZED IN AMMONIUM SULFATE 3bkc 
Crystal structure of anti-amyloid beta FAB WO2 (P21, FormB) 3bkj 
Crystal structure of Fab wo2 bound to the n terminal domain of amyloid beta peptide (1-16) 3bkm 
Structure of anti-amyloid-beta Fab WO2 (Form A, P212121) 3bky 
Crystal Structure of Chimeric Antibody C2H7 Fab in complex with a CD20 Peptide 3bn9 
Crystal Structure of MT-SP1 in complex with Fab Inhibitor E2 3bp5 
Crystal structure of the mouse PD-1 and PD-L2 complex 3bp6 
Crystal structure of the mouse PD-1 Mutant and PD-L2 complex 3bpc 
co-crystal structure of S25-2 Fab in complex with 5-deoxy-4-epi-2,3-dehydro Kdo (4.8) Kdo 3bqu 
Crystal Structure of the 2F5 Fab'-3H6 Fab Complex 3bsz 
Crystal structure of the transthyretin-retinol binding protein-Fab complex 3bt2 
Structure of urokinase receptor, urokinase and vitronectin complex 3bz4 
Crystal structure of Fab F22-4 in complex with a Shigella flexneri 2a O-Ag decasaccharide 3c08 
Crystal structure the Fab fragment of matuzumab/EMD72000 (Fab72000) 3c09 
Crystal structure the Fab fragment of matuzumab (Fab72000) in complex with domain III of the extracellular region of EGFR 3c2a 
Antibody Fab fragment 447-52D in complex with UG1033 peptide 3c2s 
Structural Characterization of a Human Fc Fragment Engineered for Lack of Effector Functions 3c5s 
Crystal Structure of monoclonal Fab F22-4 specific for Shigella flexneri 2a O-Ag 3c5z 
Crystal structure of mouse MHC class II I-Ab/3K peptide complexed with mouse TCR B3K506 3c60 
Crystal structure of mouse MHC class II I-Ab/3K peptide complexed with mouse TCR YAe62 3c6l 
Crystal structure of mouse MHC class II I-Ab/3K peptide complexed with mouse TCR 2W20 3c6s 
Crystal structure of Fab F22-4 in complex with a Shigella flexneri 2a O-Ag pentadecasaccharide 3cd4 
REFINEMENT AND ANALYSIS OF THE FIRST TWO DOMAINS OF HUMAN CD4 3cfb 
High-resolution structure of blue fluorescent antibody EP2-19G2 in complex with stilbene hapten at 100K 3cfc 
High-resolution structure of blue fluorescent antibody EP2-19G2 3cfd 
Purple-fluorescent antibody EP2-25C10 in complex with its stilbene hapten 3cfe 
Crystal structure of purple-fluorescent antibody EP2-25C10 3cfj 
Crystal structure of catalytic elimination antibody 34E4, orthorhombic crystal form 3cfk 
Crystal structure of catalytic elimination antibody 34E4, triclinic crystal form 3chn 
Solution structure of human secretory IgA1 3ck0 
ANTI-ANTI-IDIOTYPIC ANTIBODY AGAINST HUMAN ANGIOTENSIN II, COMPLEX WITH HUMAN ANGIOTENSIN II 3cle 
HIV neutralizing monoclonal antibody YZ23 3clf 
HIV neutralizing monoclonal antibody YZ23 3cm9 
Solution Structure of Human SIgA2 3cmo 
HIV neutralizing monoclonal antibody YZ18 3csy 
Crystal structure of the trimeric prefusion Ebola virus glycoprotein in complex with a neutralizing antibody from a human survivor 3cvh 
How TCR-like antibody recognizes MHC-bound peptide 3cvi 
How TCR-like antibody recognizes MHC-bound peptide 3cxd 
Crystal structure of anti-osteopontin antibody 23C3 in complex with its epitope peptide 3d0l 
Crystal structure of the HIV-1 broadly neutralizing antibody 2F5 in complex with the gp41 FP-MPER Hyb3K construct 514GIGALFLGFLGAAGS528KK-Ahx-655KNEQELLELDKWASLWN671 3d0v 
Crystal structure of the HIV-1 Cross Neutralizing Monoclonal Antibody 2F5 in complex with gp41 Peptide LLELDKWASLW 3d2u 
Structure of UL18, a Peptide-Binding Viral MHC Mimic, Bound to a Host Inhibitory Receptor 3d39 
The complex between TCR A6 and human Class I MHC HLA-A2 with the modified HTLV-1 TAX (Y5(4-fluoroPhenylalanine)) peptide 3d3v 
The complex between TCR A6 and human Class I MHC HLA-A2 with the modified HTLV-1 TAX (Y5(3,4-difluoroPhenylalanine)) peptide 3d69 
Crystal Structure of the Fab Fragment of an Anti-Factor IX Antibody 10C12 3d6g 
Fc fragment of IgG1 (Herceptin) with protein-A mimetic peptide dendrimer ligand. 3d85 
Crystal structure of IL-23 in complex with neutralizing FAB 3d9a 
High Resolution Crystal Structure Structure of HyHel10 Fab Complexed to Hen Egg Lysozyme 3det 
Structure of the E148A, Y445A doubly ungated mutant of E.coli CLC_Ec1, Cl-/H+ antiporter 3dgg 
Crystal structure of FabOX108 3dif 
Crystal structure of FabOX117 3dj9 
Crytal Structure of an isolated, unglycosylated antibody CH2 domain 3dmk 
Crystal structure of Down Syndrome Cell Adhesion Molecule (DSCAM) isoform 1.30.30, N-terminal eight Ig domains 3dnk 
Enzyme deglycosylated Human IgG1 Fc fragment 3do3 
Human 1gG1 Fc fragment, 2.5 Angstrom structure 3dro 
Crystal structure of the HIV-1 Cross Neutralizing Antibody 2F5 in complex with gp41 Peptide ELLELDKWASLWN grown in ammonium sulfate 3drq 
Crystal structure of the HIV-1 broadly neutralizing antibody 2F5 in complex with the gp41 FP-MPER Hyb3K construct 514GIGALFLGFLGAAGS528KK-Ahx-655KNEQELLELDKWASLWN671 soaked in PEG/2-propanol solution 3drt 
Crystal structure of the HIV-1 broadly neutralizing antibody 2F5 in complex with the gp41 scrambledFP-MPER scrHyb3K construct GIGAFGLLGFLAAGSKK-Ahx-K656NEQELLELDKWASLWN671 3dsf 
Crystal structure of anti-osteopontin antibody 23C3 in complex with W43A mutated epitope peptide 3dvg 
Crystal structure of K63-specific fab Apu.3A8 bound to K63-linked di-ubiquitin 3dvn 
Crystal structure of K63-specific fab Apu2.16 bound to K63-linked di-ubiquitin 3dx9 
Crystal Structure of the DM1 TCR at 2.75A 3dxa 
Crystal Structure of the DM1 TCR in complex with HLA-B*4405 and decamer EBV antigen 3e8u 
Crystal structure and thermodynamic analysis of diagnostic Fab 106.3 complexed with BNP 5-13 (C10A) reveal basis of selective molecular recognition 3efd 
The crystal structure of the cytoplasmic domain of KcsA 3eff 
The Crystal Structure of Full-Length KcsA in its Closed Conformation 3egs 
Crystal structure of the HIV-1 broadly neutralizing antibody 2F5 in complex with the gp41 scrambledFP-MPER scrHyb3K construct GIGAFGLLGFLAAGSKK-Ahx-K656NEQELLELDKWASLWN671 soaked in ammonium sulfate 3ejj 
Structure of M-CSF bound to the first three domains of FMS 3ejy 
Structure of E203H mutant of E.coli Cl-/H+ antiporter, CLC-ec1 3ejz 
Structure of E203V mutant E.coli Cl-/H+ exchanger, CLC-ec1 3eo0 
Structure of the Transforming Growth Factor-Beta Neutralizing Antibody GC-1008 3eo1 
Structure of the Fab Fragment of GC-1008 in Complex with Transforming Growth Factor-Beta 3 3eo9 
Crystal structure the Fab fragment of Efalizumab 3eoa 
Crystal structure the Fab fragment of Efalizumab in complex with LFA-1 I domain, Form I 3eob 
Crystal structure the Fab fragment of Efalizumab in complex with LFA-1 I domain, Form II 3eot 
Crystal structure of LAC031, an engineered anti-VLA1 Fab 3eyf 
Crystal structure of anti-human cytomegalovirus antibody 8f9 plus gB peptide 3eyo 
Crystal structure of anti-human cytomegalovirus antibody 8F9 3eyq 
Crystal structure of MJ5 Fab, a germline antibody variant of anti-human cytomegalovirus antibody 8f9 3eys 
PFA1 Fab fragment complexed with pyro-Glu3-A-Beta (3-8) 3eyu 
PFA1 Fab fragment complexed with Ror2(518-525) 3eyv 
Anti-Lewis Y Fab fragment with Lewis Y antigen in the presence of zinc ions 3f12 
Germline V-genes sculpt the binding site of a family of antibodies neutralizing human cytomegalovirus 3f58 
IGG1 FAB FRAGMENT (58.2) COMPLEX WITH 12-RESIDUE CYCLIC PEPTIDE (INCLUDING RESIDUES 315-324 OF HIV-1 GP120 (MN ISOLATE); H315S MUTATION 3f5w 
KcsA Potassium channel in the open-inactivated state with 32 A opening at T112 3f7v 
KcsA Potassium channel in the open-inactivated state with 23 A opening at T112 3f7y 
KcsA Potassium channel in the partially open state with 17 A opening at T112 3f8u 
Tapasin/ERp57 heterodimer 3fb5 
KcsA potassium channel in the partially open state with 14.5 A opening at T112 3fb6 
KcsA Potassium channel in the partially open state with 16 A opening at T112 3fb7 
Open KcsA potassium channel in the presence of Rb+ ion 3fb8 
KcsA Potassium channel in the open-conductive state with 20 A opening at T112 in the presence of Rb+ ion 3fct 
MATURE METAL CHELATASE CATALYTIC ANTIBODY WITH HAPTEN 3ffc 
Crystal Structure of CF34 TCR in complex with HLA-B8/FLR 3ffd 
Structure of parathyroid hormone-related protein complexed to a neutralizing monoclonal antibody 3fjt 
Crystal structure of a human Fc fragment engineered for extended serum half-life 3fmg 
Structure of rotavirus outer capsid protein VP7 trimer in complex with a neutralizing Fab 3fn0 
Crystal structure of HIV-1 neutralizing human Fab Z13e1 in complex with a 12-residue peptide containing the Z13e1 epitope on gp41 3fn3 
Dimeric Structure of PD-L1 3fo0 
Crystal structure of hapten complex of catalytic elimination antibody 13G5 (wild-type) 3fo1 
Crystal structure of hapten complex of catalytic elimination antibody 13G5 (Glu(L39)Ala mutant) 3fo2 
Crystal structure of hapten complex of catalytic elimination antibody 13G5 (Glu(L39)Gln mutant) 3fo9 
Crystal structure of aldolase antibody 33F12 Fab' in complex with hapten 1,3-diketone 3fzu 
IgG1 Fab characterized by H/D exchange 3g04 
Crystal structure of the TSH receptor in complex with a thyroid-stimulating autoantibody 3g5v 
Antibodies Specifically Targeting a Locally Misfolded Region of Tumor Associated EGFR 3g5x 
Antibodies Specifically Targeting a Locally Misfolded Region of Tumor Associated EGFR 3g5y 
Antibodies Specifically Targeting a Locally Misfolded Region of Tumor Associated EGFR 3g5z 
Antibodies Specifically Targeting a Locally Misfolded Region of Tumor Associated EGFR 3g6a 
Crystal structure of anti-IL-13 antibody CNTO607 3g6d 
Crystal structure of the complex between CNTO607 Fab and IL-13 3g6j 
C3b in complex with a C3b specific Fab 3gb7 
Potassium Channel KcsA-Fab complex in Li+ 3gbm 
Crystal Structure of Fab CR6261 in Complex with a H5N1 influenza virus hemagglutinin. 3gbn 
Crystal Structure of Fab CR6261 in Complex with the 1918 H1N1 influenza virus hemagglutinin 3ggw 
Crystal Structure of FAB F22-4 in complex with a Carbohydrate-mimetic peptide 3ghb 
Crystal structure of anti-HIV-1 Fab 447-52D in complex with V3 peptide W2RW020 3ghe 
Crystal structure of anti-HIV-1 Fab 537-10D in complex with V3 peptide MN 3gi8 
Crystal Structure of ApcT K158A Transporter Bound to 7F11 Monoclonal Fab Fragment 3gi9 
Crystal Structure of ApcT Transporter Bound to 7F11 Monoclonal Fab Fragment 3giz 
Crystal structure of the Fab fragment of anti-CD20 antibody Ofatumumab 3gje 
Rational development of high-affinity T-cell receptor-like antibodies 3gjf 
Rational development of high-affinity T-cell receptor-like antibodies 3gk8 
X-ray crystal structure of the Fab from MAb 14, mouse antibody against Canine Parvovirus 3gkw 
Crystal structure of the Fab fragment of Nimotuzumab. An anti-epidermal growth factor receptor antibody 3gnm 
The crystal structure of the JAA-F11 monoclonal antibody Fab fragment 3go1 
Crystal structure of anti-HIV-1 Fab 268-D in complex with V3 peptide MN 3grw 
FGFR3 in complex with a Fab 3gsn 
Crystal structure of the public RA14 TCR in complex with the HCMV dominant NLV/HLA-A2 epitope 3h0t 
Hepcidin-Fab complex 3h42 
Crystal structure of PCSK9 in complex with Fab from LDLR competitive antibody 3h8n 
Crystal Structure Analysis of KIR2DS4 3h9s 
The complex between TCR A6 and human Class I MHC HLA-A2 with the bound Tel1p peptide 3hae 
Rational development of high-affinity T-cell receptor-like antibodies 3hc0 
BHA10 IgG1 wild-type Fab - antibody directed at human LTBR 3hc3 
BHA10 IgG1 Fab double mutant variant - antibody directed at human LTBR 3hc4 
BHA10 IgG1 Fab quadruple mutant variant - antibody directed at human LTBR 3he6 
Crystal structure of mouse CD1d-alpha-galactosylceramide with mouse Valpha14-Vbeta8.2 NKT TCR 3he7 
Crystal structure of mouse CD1d-alpha-galactosylceramide with mouse Valpha14-Vbeta7 NKT TCR 3hfm 
STRUCTURE OF AN ANTIBODY-ANTIGEN COMPLEX. CRYSTAL STRUCTURE OF THE HY/HEL-10 FAB-LYSOZYME COMPLEX 3hg1 
Germline-governed recognition of a cancer epitope by an immunodominant human T cell receptor 3hi1 
Structure of HIV-1 gp120 (core with V3) in Complex with CD4-Binding-Site Antibody F105 3hi5 
Crystal structure of Fab fragment of AL-57 3hi6 
Crystal structure of intermediate affinity I domain of integrin LFA-1 with the Fab fragment of its antibody AL-57 3hmw 
Crystal structure of ustekinumab FAB 3hmx 
Crystal structure of ustekinumab FAB/IL-12 complex 3hns 
CS-35 Fab Complex with Oligoarabinofuranosyl Hexasaccharide 3hnt 
CS-35 Fab complex with a linear, terminal oligoarabinofuranosyl tetrasaccharide from lipoarabinomannan 3hnv 
CS-35 Fab Complex with Oligoarabinofuranosyl Tetrasaccharide (branch part of Hexasaccharide) 3hpl 
KcsA E71H-F103A mutant in the closed state 3hr5 
M1prime peptide from IgE bound by humanized antibody 47H4 Fab 3huj 
Crystal structure of human CD1d-alpha-Galactosylceramide in complex with semi-invariant NKT cell receptor 3hzk 
Crystal structure of S73-2 antibody in complex with antigen Kdo(2.4)Kdo 3hzm 
Crystal structure of S73-2 antibody in complex with antigen Kdo 3hzv 
Crystal structure of S73-2 antibody in complex with antigen Kdo(2.8)Kdo(2.4)Kdo 3hzy 
Crystal structure of S73-2 antibody in complex with antigen Kdo(2.4)Kdo(2.4)Kdo 3i02 
Crystal structure of S54-10 antibody in complex with antigen Kdo(2.4)Kdo(2.4)Kdo 3i2c 
Crystal structure of anti-IL-23 antibody CNTO4088 3i50 
Crystal structure of the West Nile Virus envelope glycoprotein in complex with the E53 antibody Fab 3i75 
Antibody Structure 3i9g 
Crystal structure of the LT1009 (SONEPCIZUMAB) antibody Fab fragment in complex with sphingosine-1-phosphate 3idg 
Crystal structure of the HIV-1 Cross Neutralizing Monoclonal Antibody 2F5 in complex with gp41 Peptide ALDKWD 3idi 
Crystal structure of the HIV-1 Cross Neutralizing Monoclonal Antibody 2F5 Fab' fragment in complex with gp41 Peptide ALDKWNQ 3idj 
Crystal structure of the HIV-1 Cross Neutralizing Monoclonal Antibody 2F5 Fab' fragment in complex with gp41 Peptide analog ELD(Orn)WAS 3idm 
Crystal structure of the HIV-1 Cross Neutralizing Monoclonal Antibody 2F5 Fab' fragment in complex with gp41 Peptide analog ELD(Nrg)WAS 3idn 
Crystal structure of the HIV-1 Cross Neutralizing Monoclonal Antibody 2F5 Fab' fragment in complex with gp41 Peptide analog ELD(Paf)WAS 3idx 
Crystal structure of HIV-gp120 core in complex with CD4-binding site antibody b13, space group C222 3idy 
Crystal structure of HIV-gp120 core in complex with CD4-binding site antibody b13, space group C2221 3iet 
Crystal Structure of 237mAb with antigen 3if1 
Crystal structure of 237mAb in complex with a GalNAc 3ifl 
X-ray structure of amyloid beta peptide:antibody (Abeta1-7:12A11) complex 3ifn 
X-ray structure of amyloid beta peptide:antibody (Abeta1-40:12A11) complex 3ifo 
X-ray structure of amyloid beta peptide:antibody (Abeta1-7:10D5) complex 3ifp 
X-ray structure of amyloid beta peptide:antibody (Abeta1-7:12B4) complex 3iga 
Potassium Channel KcsA-Fab complex in Li+ and K+ 3ijh 
Structure of S67-27 in Complex with Ko 3ijs 
Structure of S67-27 in Complex with TSBP 3ijy 
Structure of S67-27 in Complex with Kdo(2.8)Kdo 3ikc 
Structure of S67-27 in Complex with Kdo(2.8)-7-O-methyl-Kdo 3inu 
Crystal structure of an unbound KZ52 neutralizing anti-Ebolavirus antibody. 3it8 
Crystal structure of TNF alpha complexed with a poxvirus MHC-related TNF binding protein 3iu3 
Crystal structure of the Fab fragment of therapeutic antibody Basiliximab in complex with IL-2Ra (CD25) ectodomain 3iu4 
anti NeuGcGM3 ganglioside chimeric antibody chP3 3ivk 
Crystal Structure of the Catalytic Core of an RNA Polymerase Ribozyme Complexed with an Antigen Binding Antibody Fragment 3ixt 
Crystal Structure of Motavizumab Fab Bound to Peptide Epitope 3ixx 
The pseudo-atomic structure of West Nile immature virus in complex with Fab fragments of the anti-fusion loop antibody E53 3ixy 
The pseudo-atomic structure of dengue immature virus in complex with Fab fragments of the anti-fusion loop antibody E53 3iyw 
West Nile virus in complex with Fab fragments of MAb CR4354 (fitted coordinates of envelope proteins and Fab fragments of one icosahedral ASU) 3j1s 
Structure of adeno-associated virus-2 in complex with neutralizing monoclonal antibody A20 3j2x 
Electron Cryo-microscopy of Chikungunya VLP in complex with neutralizing antibody Fab m242 3j2y 
Electron Cryo-microscopy of Chikungunya VLP in complex with neutralizing antibody Fab 9.8B 3j2z 
Electron Cryo-microscopy of Chikungunya VLP in complex with neutralizing antibody Fab m10 3j30 
Electron Cryo-microscopy of Chikungunya VLP in complex with neutralizing antibody Fab CHK152 3j3o 
Conformational Shift of a Major Poliovirus Antigen Confirmed by Immuno-Cryogenic Electron Microscopy: 160S Poliovirus and C3-Fab Complex 3j3p 
Conformational Shift of a Major Poliovirus Antigen Confirmed by Immuno-Cryogenic Electron Microscopy: 135S Poliovirus and C3-Fab Complex 3j3z 
Structure of MA28-7 neutralizing antibody Fab fragment from electron cryo-microscopy of enterovirus 71 complexed with a Fab fragment 3j42 
Obstruction of Dengue Virus Maturation by Fab Fragments of the 2H2 Antibody 3j5m 
Cryo-EM structure of the BG505 SOSIP.664 HIV-1 Env trimer with 3 PGV04 Fabs 3j6n 
Kinetic and Structural Analysis of Coxsackievirus B3 Receptor Interactions and Formation of the A-particle 3j6o 
Kinetic and Structural Analysis of Coxsackievirus B3 Receptor Interactions and Formation of the A-particle 3j70 
3J70 3j8d 
3J8D 3j8f 
3J8F 3j93 
3J93 3jab 
3JAB 3jbq 
3JBQ 3jwd 
Structure of HIV-1 gp120 with gp41-Interactive Region: Layered Architecture and Basis of Conformational Mobility 3jwo 
Structure of HIV-1 gp120 with gp41-Interactive Region: Layered Architecture and Basis of Conformational Mobility 3jxa 
Immunoglobulin domains 1-4 of mouse CNTN4 3jz7 
Crystal structure of the extracellular domains of coxsackie & adenovirus receptor from mouse (mCAR) 3k0w 
Crystal structure of the tandem IG-like C2-type 2 domains of the human mucosa-associated lymphoid tissue lymphoma translocation protein 1 3k2u 
Crystal structure of HGFA in complex with the allosteric inhibitory antibody Fab40 3kdm 
Crystal Structure of Human Anti-steroid Fab 5F2 in Complex with Testosterone 3kho 
Crystal structure of murine Ig-beta (CD79b) homodimer 3khq 
Crystal structure of murine Ig-beta (CD79b) in the monomeric form 3kj4 
Structure of rat Nogo receptor bound to 1D9 antagonist antibody 3kj6 
Crystal structure of a Methylated beta2 Adrenergic Receptor-Fab complex 3kld 
PTPRG CNTN4 complex 3klh 
Crystal structure of AZT-Resistant HIV-1 Reverse Transcriptase crosslinked to post-translocation AZTMP-Terminated DNA (COMPLEX P) 3kpr 
Crystal Structure of the LC13 TCR in complex with HLA B*4405 bound to EEYLKAWTF a mimotope 3kps 
Crystal Structure of the LC13 TCR in complex with HLA B*4405 bound to EEYLQAFTY a self peptide from the ABCD3 protein 3kr3 
Crystal structure of IGF-II antibody complex 3ks0 
Crystal structure of the heme domain of flavocytochrome b2 in complex with Fab B2B4 3kxf 
Crystal Structure of SB27 TCR in complex with the 'restriction triad' mutant HLA-B*3508-13mer 3kyk 
Crystal structure of li33 Igg1 Fab 3kym 
Crystal structure of Li33 IgG2 di-Fab 3l1o 
Crystal structure of monoclonal antibody MN423 Fab fragment with free combining site, crystallized in the presence of zinc 3l5w 
Crystal structure of the complex between IL-13 and C836 FAB 3l5x 
Crystal structure of the complex between IL-13 and H2L6 FAB 3l7e 
Crystal structure of ANTI-IL-13 antibody C836 3l7f 
Structure of IL-13 antibody H2L6, A humanized variant OF C836 3l95 
Crystal structure of the human Notch1 Negative Regulatory Region (NRR) bound to the fab fragment of an antagonist antibody 3laf 
Structure of DCC, a netrin-1 receptor 3lcy 
Titin Ig tandem domains A164-A165 3ld8 
Structure of JMJD6 and Fab Fragments 3ldb 
Structure of JMJD6 complexd with ALPHA-KETOGLUTARATE and Fab Fragment. 3lev 
HIV-1 antibody 2F5 in complex with epitope scaffold ES2 3lex 
2F5 Epitope scaffold elicited anti-HIV-1 monoclonal antibody 11F10 in complex with HIV-1 GP41 3ley 
2F5 Epitope scaffold elicited anti-HIV-1 monoclonal antibody 6a7 in complex with HIV-1 GP41 3liz 
crystal structure of bla g 2 complexed with Fab 4C3 3lmj 
Structure of human anti HIV 21c Fab 3lqa 
Crystal structure of clade C gp120 in complex with sCD4 and 21c Fab 3lrs 
Structure of PG16, an antibody with broad and potent neutralization of HIV-1 3ls4 
Crystal Structure of Anti-tetrahydrocannabinol Fab Fragment in Complex with THC 3ls5 
Anti-tetrahydrocannabinol Fab Fragment, Free Form 3lzf 
Crystal Structure of Fab 2D1 in Complex with the 1918 Influenza Virus Hemagglutinin 3m8o 
Human IgA1 Fab fragment 3ma9 
Crystal structure of gp41 derived protein complexed with fab 8066 3mac 
crystal structure of GP41-derived protein complexed with fab 8062 3mbe 
TCR 21.30 in complex with MHC class II I-Ag7HEL(11-27) 3mbx 
Crystal structure of chimeric antibody X836 3mcg 
THREE-DIMENSIONAL STRUCTURE OF A LIGHT CHAIN DIMER CRYSTALLIZED IN WATER. CONFORMATIONAL FLEXIBILITY OF A MOLECULE IN TWO CRYSTAL FORMS 3mck 
Crystal structure of anti-beta-amyloid antibody C705 3mcl 
Anti-beta-amyloid antibody c706 fab in space group P21 3mff 
1F1E8hu TCR 3mj7 
Crystal structure of the complex of JAML and Coxsackie and Adenovirus receptor, CAR 3mj8 
Crystal structure of HL4E10 Fab, a hamster Ab stimulatory for gammadelta T cells 3mj9 
Crystal structure of JAML in complex with the stimulatory antibody HL4E10 3mjg 
The structure of a platelet derived growth factor receptor complex 3mlr 
Crystal structure of anti-HIV-1 V3 Fab 2557 in complex with a NY5 V3 peptide 3mls 
Crystal structure of anti-HIV-1 V3 mAb 2557 Fab in complex with a HIV-1 gp120 V3 mimotope 3mlt 
Crystal structure of anti-HIV-1 V3 Fab 2557 in complex with a UG1033 V3 peptide 3mlu 
Crystal structure of anti-HIV-1 V3 Fab 2557 in complex with a ZAM18 V3 peptide 3mlv 
Crystal structure of anti-HIV-1 V3 Fab 2557 in complex with an NOF V3 peptide 3mlw 
Crystal structure of anti-HIV-1 V3 Fab 1006-15D in complex with an MN V3 peptide 3mlx 
Crystal structure of anti-HIV-1 V3 Fab 3074 in complex with an MN V3 peptide 3mly 
Crystal structure of anti-HIV-1 V3 Fab 3074 in complex with a UR29 V3 peptide 3mlz 
Crystal structure of anti-HIV-1 V3 Fab 3074 in complex with a VI191 V3 peptide 3mme 
Structure and functional dissection of PG16, an antibody with broad and potent neutralization of HIV-1 3mnv 
Crystal structure of the non-neutralizing HIV antibody 13H11 Fab fragment 3mnw 
Crystal structure of the non-neutralizing HIV antibody 13H11 Fab fragment with a gp41 MPER-derived peptide in a helical conformation 3mnz 
Crystal structure of the non-neutralizing HIV antibody 13H11 Fab fragment with a gp41 MPER-derived peptide bearing Ala substitutions in a helical conformation 3mo1 
Crystal structure of the non-neutralizing HIV antibody 13H11 Fab fragment 3moa 
Crystal structure of the neutralizing HIV antibody 2F5 Fab fragment (recombinantly produced Fab) with 17 aa gp41 MPER-derived peptide 3mob 
Crystal structure of the neutralizing HIV antibody 2F5 Fab fragment (recombinantly produced Fab) with 11 aa gp41 MPER-derived peptide 3mod 
Crystal structure of the neutralizing HIV antibody 2F5 Fab fragment (recombinantly produced IgG) with 11 aa gp41 MPER-derived peptide 3mug 
Crystal structure of human Fab PG16, a broadly reactive and potent HIV-1 neutralizing antibody 3muh 
Crystal structure of PG9 light chain 3mv7 
Crystal Structure of the TK3 TCR in complex with HLA-B*3501/HPVG 3mv8 
Crystal Structure of the TK3-Gln55His TCR in complex with HLA-B*3501/HPVG 3mv9 
Crystal Structure of the TK3-Gln55Ala TCR in complex with HLA-B*3501/HPVG 3mxv 
Crystal structure of fab fragment of anti-Shh 5E1 chimera 3mxw 
Crystal structure Sonic hedgehog bound to the 5E1 fab fragment 3n85 
Crystallographic trimer of HER2 extracellular regions in complex with tryptophan-rich antibody fragment 3n9g 
Crystal structure of the Fab fragment of the human neutralizing anti-West Nile Virus MAb CR4354 3na9 
Crystal structure of Fab15 3naa 
Crystal structure of Fab15 Mut5 3nab 
Crystal Structure of fab15 Mut6 3nac 
Crystal structure of Fab15 Mut7 3ncj 
Crystal structure of Fab15 Mut8 3ncy 
X-ray crystal structure of an arginine agmatine antiporter (AdiC) in complex with a Fab fragment 3nfp 
Crystal structure of the Fab fragment of therapeutic antibody daclizumab in complex with IL-2Ra (CD25) ectodomain 3nfs 
Crystal structure the Fab fragment of therapeutic antibody daclizumab 3ngb 
Crystal structure of broadly and potently neutralizing antibody VRC01 in complex with HIV-1 gp120 3nh7 
Crystal structure of the neutralizing Fab fragment AbD1556 bound to the BMP type I receptor IA 3nid 
The Closed Headpiece of Integrin alphaIIB beta3 and its Complex with an alpahIIB beta3 -Specific Antagonist That Does Not Induce Opening 3nif 
The Closed Headpiece of Integrin IIb 3 and its Complex with an IIb 3 -Specific Antagonist That Does Not Induce Opening 3nig 
The Closed Headpiece of Integrin IIb 3 and its Complex with an IIb 3 -Specific Antagonist That Does Not Induce Opening 3nps 
Crystal structure of membrane-type serine protease 1 (MT-SP1) in complex with the Fab Inhibitor S4 3ntc 
Crystal structure of KD-247 Fab, an anti-V3 antibody that inhibits HIV-1 Entry 3nz8 
Crystal structure of the HIV-2 neutralizing Fab fragment 7C8 3nzh 
Crystal structure of anti-emmprin antibody 5F6 FAB 3o0r 
Crystal structure of nitric oxide reductase from Pseudomonas aeruginosa in complex with antibody fragment 3o11 
Anti-beta-amyloid antibody c706 fab in space group c2 3o2d 
Crystal structure of HIV-1 primary receptor CD4 in complex with a potent antiviral antibody 3o2v 
Crystal structure of 1E9 PheL89Ser/LeuH47Trp/MetH100bPhe, an engineered Diels-Alderase Fab with modified specificity and catalytic activity 3o2w 
Crystal structure of the 1E9 PheL89Ser/LeuH47Trp/MetH100bPhe Fab in complex with a 39A11 transition state analog 3o41 
Crystal Structure of 101F Fab Bound to 15-mer Peptide Epitope 3o45 
Crystal Structure of 101F Fab Bound to 17-mer Peptide Epitope 3o4l 
Genetic and structural basis for selection of a ubiquitous T cell receptor deployed in Epstein-Barr virus 3o6f 
Crystal structure of a human autoimmune TCR MS2-3C8 bound to MHC class II self-ligand MBP/HLA-DR4 3o6k 
Crystal structure of anti-Tat HIV Fab'11H6H1 3o6l 
Anti-Tat HIV 11H6H1 Fab' complexed with a 15-mer Tat peptide 3o6m 
Anti-Tat HIV 11H6H1 Fab' complexed with a 9-mer Tat peptide 3o8x 
Recognition of Glycolipid Antigen by iNKT Cell TCR 3o9w 
Recognition of a Glycolipid Antigen by the iNKT Cell TCR 3oau 
Antibody 2G12 Recognizes Di-Mannose Equivalently in Domain- and Non-Domain-Exchanged Forms, but only binds the HIV-1 Glycan Shield if Domain-Exchanged 3oay 
A non-self sugar mimic of the HIV glycan shield shows enhanced antigenicity 3oaz 
A non-self sugar mimic of the HIV glycan shield shows enhanced antigenicity 3ob0 
A non-self sugar mimic of the HIV glycan shield shows enhanced antigenicity 3of6 
Human pre-T cell receptor crystal structure 3ogc 
KcsA E71A variant in presence of Na+ 3ojd 
Anti-Indolicidin monoclonal antibody V2D2 (Fab fragment) 3okd 
Crystal structure of S25-39 in complex with Kdo 3oke 
Crystal structure of S25-39 in complex with Ko 3okk 
Crystal structure of S25-39 in complex with Kdo(2.4)Kdo 3okl 
Crystal structure of S25-39 in complex with Kdo(2.8)Kdo 3okm 
Crystal structure of unliganded S25-39 3okn 
Crystal structure of S25-39 in complex with Kdo(2.4)Kdo(2.4)Kdo 3oko 
Crystal structure of S25-39 in complex with Kdo(2.8)Kdo(2.4)Kdo 3omz 
Crystal structure of MICA-specific human gamma delta T cell receptor 3opz 
Crystal structure of trans-sialidase in complex with the Fab fragment of a neutralizing monoclonal IgG antibody 3oq3 
Structural Basis of Type-I Interferon Sequestration by a Poxvirus Decoy Receptor 3or6 
On the structural basis of modal gating behavior in K+channels - E71Q 3or7 
On the structural basis of modal gating behavior in K+channels - E71I 3oz9 
Crystal Structure of anti-gp41 Fab NC-1 3p0v 
anti-EGFR/HER3 Fab DL11 alone 3p0y 
anti-EGFR/HER3 Fab DL11 in complex with domain III of EGFR extracellular region 3p11 
anti-EGFR/HER3 Fab DL11 in complex with domains I-III of the HER3 extracellular region 3p2t 
Crystal Structure of Leukocyte Ig-like Receptor LILRB4 (ILT3/LIR-5/CD85k) 3p30 
crystal structure of the cluster II Fab 1281 in complex with HIV-1 gp41 ectodomain 3p3y 
Crystal structure of neurofascin homophilic adhesion complex in space group p6522 3p40 
Crystal structure of neurofascin adhesion complex in space group p3221 3pgf 
Crystal structure of maltose bound MBP with a conformationally specific synthetic antigen binder (sAB) 3pho 
Crystal structure of S64-4 in complex with PSBP 3phq 
Crystal structure of S64-4 in complex with KDO 3piq 
Crystal structure of human 2909 Fab, a quaternary structure-specific antibody against HIV-1 3pjs 
Mechanism of Activation Gating in the Full-Length KcsA K+ Channel 3pl6 
Structure of Autoimmune TCR Hy.1B11 in complex with HLA-DQ1 and MBP 85-99 3pnw 
Crystal Structure of the tudor domain of human TDRD3 in complex with an anti-TDRD3 FAB 3pp3 
Epitope characterization and crystal structure of GA101 provide insights into the molecular basis for the type I / type II distinction of anti- CD20 antibodies 3pp4 
Epitope characterization and crystal structure of GA101 provide insights into the molecular basis for the type I / type II distinction of anti- CD20 antibodies 3pqy 
Crystal Structure of 6218 TCR in complex with the H2Db-PA224 3pv6 
Crystal structure of NKp30 bound to its ligand B7-H6 3pv7 
Crystal structure of NKp30 ligand B7-H6 3pwp 
The complex between TCR A6 and human Class I MHC HLA-A2 with the bound HuD peptide 3q1s 
HIV-1 neutralizing antibody Z13e1 in complex with epitope display protein 3q3g 
Crystal Structure of A-domain in complex with antibody 3q5t 
V beta/V beta homodimerization-based pre-TCR model suggested by TCR beta crystal structures 3q5y 
V beta/V beta homodimerization-based pre-TCR model suggested by TCR beta crystal structures 3q6f 
Crystal structure of Fab of human mAb 2909 specific for quaternary neutralizing epitope of HIV-1 gp120 3q6g 
Crystal structure of Fab of rhesus mAb 2.5B specific for quaternary neutralizing epitope of HIV-1 gp120 3qa3 
Crystal Structure of A-domain in complex with antibody 3qct 
Crystal structure of the humanized apo LT3015 anti-lysophosphatidic acid antibody Fab fragment 3qcu 
Crystal structure of the LT3015 antibody Fab fragment in complex with lysophosphatidic acid (14:0) 3qcv 
Crystal structure of the LT3015 antibody Fab fragment in complex with lysophosphatidic acid (18:2) 3qdg 
The complex between TCR DMF5 and human Class I MHC HLA-A2 with the bound MART-1(26-35)(A27L) peptide 3qdj 
The complex between TCR DMF5 and human Class I MHC HLA-A2 with the bound MART-1(27-35) nonameric peptide 3qdm 
The complex between TCR DMF4 and human Class I MHC HLA-A2 with the bound MART-1(26-35)(A27L) decameric peptide 3qeg 
Crystal structure of human N12-i2 Fab, an ADCC and neutralizing anti-HIV-1 Env antibody 3qeh 
Crystal structure of human N12-i15, an ADCC and non-neutralizing anti-HIV-1 Env antibody 3qeq 
The complex between TCR DMF4 and human Class I MHC HLA-A2 with the bound MART-1(27-35) nonameric peptide 3qeu 
The crystal structure of TCR DMF5 3qfj 
The complex between TCR A6 and human Class I MHC HLA-A2 with the modified TAX (Y5F) peptide 3qg6 
Structural Basis for Ligand Recognition and Discrimination of a Quorum Quenching Antibody 3qg7 
Structural Basis for Ligand Recognition and Discrimination of a Quorum Quenching Antibody 3qh3 
The crystal structure of TCR A6 3qhf 
Crystal Structure of Fab del2D1, a deletion variant of anti-influenza antibody 2D1 3qhz 
Crystal Structure of human anti-influenza Fab 2D1 3qi9 
Crystal structure of mouse CD1d-alpha-phosphotidylinositol with mouse Valpha14-Vbeta6 2A3-D NKT TCR 3qib 
Crystal structure of the 2B4 TCR in complex with MCC/I-Ek 3qiu 
Crystal structure of the 226 TCR in complex with MCC/I-Ek 3qiw 
Crystal structure of the 226 TCR in complex with MCC-p5E/I-Ek 3qjf 
Crystal structure of the 2B4 TCR 3qjh 
The crystal structure of the 5c.c7 TCR 3qnx 
Orthorhombic form of human IgA1 Fab fragment, sharing same Fv as IgG 3qny 
Monoclinic form of human IgA1 Fab fragment, sharing same Fv as IgG 3qnz 
Orthorhombic form of IgG1 Fab fragment (in complex with antigenic tubulin peptide) sharing same Fv as IgA 3qo0 
Monoclinic form of IgG1 Fab fragment (in complex with antigenic peptide) sharing same Fv as IgA 3qo1 
Monoclinic form of IgG1 Fab fragment (apo form) sharing same Fv as IgA 3qpq 
Crystal structure of ANTI-TLR3 antibody C1068 FAB 3qpx 
Crystal structure of Fab C2507 3qq9 
Crystal structure of FAB fragment of anti-human RSV (RESPIRATORY SYNCYTIAL VIRUS) F Protein MAB 101F 3qrg 
Crystal structure of antiRSVF Fab B21m 3qs7 
Crystal structure of a human Flt3 ligand-receptor ternary complex 3qs9 
Crystal structure of a human Flt3 ligand-receptor ternary complex 3qum 
Crystal structure of human prostate specific antigen (PSA) in Fab sandwich with a high affinity and a PCa selective antibody 3qux 
Structure of the mouse CD1d-alpha-C-GalCer-iNKT TCR complex 3quy 
Structure of the mouse CD1d-BnNH-GSL-1'-iNKT TCR complex 3quz 
Structure of the mouse CD1d-NU-alpha-GalCer-iNKT TCR complex 3qwo 
Crystal structure of a motavizumab epitope-scaffold bound to motavizumab Fab 3r06 
Crystal structure of anti-mouse CD3epsilon antibody 2C11 Fab fragment 3r08 
Crystal structure of mouse cd3epsilon in complex with antibody 2C11 Fab 3r1g 
Structure Basis of Allosteric Inhibition of BACE1 by an Exosite-Binding Antibody 3ra7 
Bispecific digoxigenin binding antibodies for targeted payload delivery 3raj 
Crystal structure of human CD38 in complex with the Fab fragment of antibody HB7 3rbs 
Crystal structure of the myomesin domains 10 and 11 3rdt 
Crystal Structure of 809.B5 TCR complexed with MHC Class II I-Ab/3k peptide 3rev 
Crystal structure of human alloreactive tcr nb20 3rgv 
A single TCR bound to MHCI and MHC II reveals switchable TCR conformers 3rhw 
C. elegans glutamate-gated chloride channel (GluCl) in complex with Fab and ivermectin 3ri5 
C. elegans glutamate-gated chloride channel (GluCl) in complex with Fab, ivermectin and picrotoxin 3ria 
C. elegans glutamate-gated chloride channel (GluCl) in complex with Fab, ivermectin and iodide. 3rif 
C. elegans glutamate-gated chloride channel (GluCl) in complex with Fab, ivermectin and glutamate. 3rjd 
Crystal structure of Fc RI and its implication to high affinity immunoglobulin G binding 3rkd 
Hepatitis E Virus E2s domain (Genotype I) in complex with a neutralizing antibody 3rnq 
Crystal structure of the complex between the extracellular domains of mouse PD-1 mutant and PD-L2 3rpi 
Crystal Structure of Fab from 3BNC60, Highly Potent anti-HIV Antibody 3rtq 
Structure of the mouse CD1d-HS44-iNKT TCR complex 3ru8 
Structure of an HIV epitope scaffold in complex with neutralizing antibody b12 Fab 3rug 
Crystal structure of Valpha10-Vbeta8.1 NKT TCR in complex with CD1d-alphaglucosylceramide (C20:2) 3rvt 
Structure of 4C1 Fab in P212121 space group 3rvu 
Structure of 4C1 Fab in C2221 space group 3rvv 
Crystal structure of Der f 1 complexed with Fab 4C1 3rvw 
Crystal structure of Der p 1 complexed with Fab 4C1 3rvx 
Crystal structure of Der p 1 complexed with Fab 4C1 3ry6 
Complex of fcgammariia (CD32) and the FC of human IGG1 3rzc 
Structure of the self-antigen iGb3 bound to mouse CD1d and in complex with the iNKT TCR 3s34 
Structure of the 1121B Fab fragment 3s35 
Structural basis for the function of two anti-VEGF receptor antibodies 3s36 
Structural basis for the function of two anti-VEGF receptor antibodies 3s37 
Structural basis for the function of two anti-VEGF receptor antibodies 3s4s 
Crystal structure of CD4 mutant bound to HLA-DR1 3s5l 
Crystal structure of CD4 mutant bound to HLA-DR1 3s62 
Structure of Fab fragment of malaria transmission blocking antibody 2A8 against P. vivax P25 protein 3s7g 
Aglycosylated human igg1 fc fragment 3s88 
Crystal structure of Sudan Ebolavirus Glycoprotein (strain Gulu) bound to 16F6 3s96 
Crystal structure of 3B5H10 3s97 
PTPRZ CNTN1 complex 3sbw 
Crystal structure of the complex between the extracellular domains of mouse PD-1 mutant and human PD-L1 3scm 
Crystal structure of autoreactive-Valpha14-Vbeta6 NKT TCR in complex with CD1d-isoglobotrihexosylceramide 3sda 
Crystal structure of autoreactive-Valpha14-Vbeta6 NKT TCR in complex with CD1d-beta-galactosylceramide 3sdc 
Crystal structure of autoreactive-Valpha14-Vbeta6 NKT TCR in complex with CD1d-globotrihexosylceramide 3sdd 
Crystal structure of autoreactive-Valpha14-Vbeta6 NKT TCR in complex with CD1d-beta-lactosylceramide 3sdx 
Crystal structure of human autoreactive-Valpha24 NKT TCR in complex with CD1d-beta-galactosylceramide 3sdy 
Crystal Structure of Broadly Neutralizing Antibody CR8020 Bound to the Influenza A H3 Hemagglutinin 3se8 
Crystal structure of broadly and potently neutralizing antibody VRC03 in complex with HIV-1 gp120 3se9 
Crystal structure of broadly and potently neutralizing antibody VRC-PG04 in complex with HIV-1 gp120 3sgd 
Crystal structure of the mouse mAb 17.2 3sge 
Crystal structure of mAb 17.2 in complex with R13 peptide 3sgj 
Unique carbohydrate-carbohydrate interactions are required for high affinity binding between FcgIII and antibodies lacking core fucose 3sgk 
Unique carbohydrate/carbohydrate interactions are required for high affinity binding of FcgIII and antibodies lacking core fucose 3shs 
Three N-terminal domains of the bacteriophage RB49 Highly Immunogenic Outer Capsid protein (Hoc) 3sjv 
Crystal structure of the RL42 TCR in complex with HLA-B8-FLR 3skj 
Structural And Functional Characterization of an Agonistic Anti-Human EphA2 Monoclonal Antibody 3skn 
Crystal structure of the RL42 TCR unliganded 3sku 
Herpes simplex virus glycoprotein D bound to the human receptor nectin-1 3sm5 
Influenza hemagglutinin in complex with a neutralizing antibody 3so3 
Structures of Fab-Protease Complexes Reveal a Highly Specific Non-Canonical Mechanism of Inhibition. 3sob 
The structure of the first YWTD beta propeller domain of LRP6 in complex with a FAB 3sqo 
PCSK9 J16 Fab complex 3stl 
KcsA potassium channel mutant Y82C with Cadmium bound 3stz 
KcsA potassium channel mutant Y82C with nitroxide spin label 3sy0 
S25-2- A(2-8)-A(2-4)KDO trisaccharide complex 3t0e 
Crystal structure of a complete ternary complex of T cell receptor, peptide-MHC and CD4 3t2n 
Human hepsin protease in complex with the Fab fragment of an inhibitory antibody 3t3m 
A Novel High Affinity Integrin alphaIIbbeta3 Receptor Antagonist That Unexpectedly Displaces Mg2+ from the beta3 MIDAS 3t3p 
A Novel High Affinity Integrin alphaIIbbeta3 Receptor Antagonist That Unexpectedly Displaces Mg2+ from the beta3 MIDAS 3t4y 
S25-2- KDO monosaccharide complex 3t65 
S25-2- A(2-8)KDO disaccharide complex 3t77 
S25-2- A(2-4)KDO disaccharide complex 3ta3 
Structure of the mouse CD1d-Glc-DAG-s2-iNKT TCR complex 3tcl 
Crystal Structure of HIV-1 Neutralizing Antibody CH04 3tfk 
42F3-p4B10/H2-Ld 3thm 
Crystal structure of Fas receptor extracellular domain in complex with Fab EP6b_B01 3tje 
Crystal structure of Fas receptor extracellular domain in complex with Fab E09 3tjh 
42F3-p3A1/H2-Ld complex 3tn0 
Structure of mouse Va14Vb8.2NKT TCR-mouse CD1d-a-C-Galactosylceramide complex 3tnm 
Crystal structure of A32 Fab, an ADCC mediating anti-HIV-1 antibody 3tnn 
Crystal structure of N5-i5 Fab, an ADCC mediating and non-neutralizing CD4i anti-HIV- 1 antibody. 3to4 
Structure of mouse Valpha14Vbeta2-mouseCD1d-alpha-Galactosylceramide 3tpu 
42F3 p5E8/H2-Ld complex 3tt1 
Crystal Structure of LeuT in the outward-open conformation in complex with Fab 3tt3 
Crystal Structure of LeuT in the inward-open conformation in complex with Fab 3tv3 
Crystal structure of broad and potent HIV-1 neutralizing antibody PGT128 in complex with Man9 3tvm 
Structure of the mouse CD1d-SMC124-iNKT TCR complex 3twc 
Crystal structure of broad and potent HIV-1 neutralizing antibody PGT127 in complex with Man9 3tyf 
Crystal structure of a CD1d-lysophosphatidylcholine reactive iNKT TCR 3tyg 
Crystal structure of broad and potent HIV-1 neutralizing antibody PGT128 in complex with a glycosylated engineered gp120 outer domain with miniV3 (eODmV3) 3tzv 
Crystal structure of an iNKT TCR in complex with CD1d-lysophosphatidylcholine 3u0t 
Fab-antibody complex 3u0w 
AD related murine antibody Fragment 3u1s 
Crystal structure of human Fab PGT145, a broadly reactive and potent HIV-1 neutralizing antibody 3u2s 
Crystal Structure of PG9 Fab in Complex with V1V2 Region from HIV-1 strain ZM109 3u30 
Crystal structure of a linear-specific Ubiquitin fab bound to linear ubiquitin 3u36 
Crystal Structure of PG9 Fab 3u46 
CH04H/CH02L P212121 3u4b 
CH04H/CH02L Fab P4 3u4e 
Crystal Structure of PG9 Fab in Complex with V1V2 Region from HIV-1 strain CAP45 3u7w 
Crystal structure of NIH45-46 Fab 3u7y 
Structure of NIH45-46 Fab in complex with gp120 of 93TH057 HIV 3u82 
Binding of herpes simplex virus glycoprotein D to nectin-1 exploits host cell adhesion 3u83 
Crystal structure of nectin-1 3u9p 
Crystal Structure of Murine Siderocalin in Complex with an Fab Fragment 3u9u 
Crystal Structure of Extracellular Domain of Human ErbB4/Her4 in complex with the Fab fragment of mAb1479 3uaj 
Crystal structure of the envelope glycoprotein ectodomain from dengue virus serotype 4 in complex with the fab fragment of the chimpanzee monoclonal antibody 5H2 3ubx 
Crystal structure of the mouse CD1d-C20:2-aGalCer-L363 mAb Fab complex 3uc0 
Crystal structure of domain I of the envelope glycoprotein ectodomain from dengue virus serotype 4 in complex with the fab fragment of the chimpanzee monoclonal antibody 5H2 3uez 
Crystal structure of the human Colony-Stimulating Factor 1 (hCSF-1) cytokine in complex with the viral receptor BARF1 3uji 
Crystal structure of anti-HIV-1 V3 Fab 2558 in complex with MN peptide 3ujj 
Crystal structure of anti-HIV-1 V3 Fab 4025 in complex with Con A peptide 3ujt 
Structure of the Fab fragment of Ab-52, an antibody that binds the O-antigen of Francisella tularensis 3uls 
Crystal structure of Fab12 3ulu 
Structure of quaternary complex of human TLR3ecd with three Fabs (Form1) 3ulv 
Structure of quaternary complex of human TLR3ecd with three Fabs (Form2) 3uo1 
Structure of a monoclonal antibody complexed with its MHC-I antigen 3utp 
1E6 TCR specific for HLA-A*0201-ALWGPDPAAA 3uts 
1E6-A*0201-ALWGPDPAAA Complex, Monoclinic 3utt 
1E6-A*0201-ALWGPDPAAA Complex, Triclinic 3utz 
Endogenous-like inhibitory antibodies targeting activated metalloproteinase motifs show therapeutic potential 3uyr 
Structure of a monoclonal antibody complexed with its MHC-I antigen 3v0v 
Fab WN1 222-5 unliganded 3v0w 
Crystal structure of Fab WN1 222-5 in complex with LPS 3v2a 
VEGFR-2/VEGF-A COMPLEX STRUCTURE 3v4p 
crystal structure of a4b7 headpiece complexed with Fab ACT-1 3v4u 
Structure of a monoclonal antibody complexed with its MHC-I antigen 3v4v 
crystal structure of a4b7 headpiece complexed with Fab ACT-1 and RO0505376 3v52 
Structure of a monoclonal antibody complexed with its MHC-I antigen 3v6b 
VEGFR-2/VEGF-E complex structure 3v6f 
Crystal Structure of an anti-HBV e-antigen monoclonal Fab fragment (e6), unbound 3v6o 
Leptin Receptor-antibody complex 3v6z 
Crystal Structure of Hepatitis B Virus e-antigen 3v7a 
Structural basis for broad detection of genogroup II noroviruses by a monoclonal antibody that binds to a site occluded in the viral particle 3v7m 
Crystal structure of monoclonal human anti-Rhesus D Fc IgG1 T125(YB2/0) in the presence of Zn2+ 3v8c 
Crystal structure of monoclonal human anti-rhesus D Fc IgG1 t125(yb2/0) double mutant (H310 and H435 in K) 3v95 
Crystal structure of monoclonal human anti-rhesus D Fc and IgG1 t125(yb2/0) in the presence of EDTA 3ve0 
Crystal structure of Sudan Ebolavirus Glycoprotein (strain Boniface) bound to 16F6 3vfg 
Crystal structure of monoclonal anitbody 3F8 Fab fragment that binds to GD2 ganglioside 3vg0 
Antibody Fab fragment 3vg9 
Crystal structure of human adenosine A2A receptor with an allosteric inverse-agonist antibody at 2.7 A resolution 3vga 
Crystal structure of human adenosine A2A receptor with an allosteric inverse-agonist antibody at 3.1 A resolution 3vi3 
Crystal structure of alpha5beta1 integrin headpiece (ligand-free form) 3vi4 
Crystal structure of alpha5beta1 integrin headpiece in complex with RGD peptide 3vrl 
Crystal structure of BMJ4 p24 capsid protein in complex with A10F9 Fab 3vw3 
Antibody 64M-5 Fab in complex with a double-stranded DNA (6-4) photoproduct 3vwj 
Ternary crystal structure of the human NKT TCR-CD1d-C20:2 complex 3vwk 
Ternary crystal structure of the human NKT TCR-CD1d-4'deoxy-alpha-galactosylceramide complex 3vxm 
The complex between C1-28 TCR and HLA-A24 bound to HIV-1 Nef134-10(2F) peptide 3vxq 
H27-14 TCR specific for HLA-A24-Nef134-10 3vxr 
The complex between H27-14 TCR and HLA-A24 bound to HIV-1 Nef134-10(wt) peptide 3vxs 
The complex between H27-14 TCR and HLA-A24 bound to HIV-1 Nef134-10(6L) peptide 3vxt 
T36-5 TCR specific for HLA-A24-Nef134-10 3vxu 
The complex between T36-5 TCR and HLA-A24 bound to HIV-1 Nef134-10(2F) peptide 3w0w 
The complex between T36-5 TCR and HLA-A24 bound to HIV-1 Nef134-10(2F) peptide in space group P212121 3w14 
Insulin receptor ectodomain construct comprising domains L1,CR,L2, FNIII-1 and alphact peptide in complex with bovine insulin and FAB 83-14 3w2d 
Crystal Structure of Staphylococcal Eenterotoxin B in complex with a novel neutralization monoclonal antibody Fab fragment 3w9d 
Structure of Human Monoclonal Antibody E317 Fab 3w9e 
Structure of Human Monoclonal Antibody E317 Fab Complex with HSV-2 gD 3wd5 
Crystal structure of TNFalpha in complex with Adalimumab Fab fragment 3we6 
3WE6 3wfb 
3WFB 3wfc 
3WFC 3wfd 
3WFD 3wfe 
3WFE 3wfh 
3WFH 3whe 
A new conserved neutralizing epitope at the globular head of hemagglutinin in H3N2 influenza viruses 3whx 
3WHX 3wif 
3WIF 3wih 
3WIH 3wii 
3WII 3wjj 
Crystal structure of IIb selective Fc variant, Fc(P238D), in complex with FcgRIIb 3wjl 
Crystal structure of IIb selective Fc variant, Fc(V12), in complex with FcgRIIb 3wkm 
The periplasmic PDZ tandem fragment of the RseP homologue from Aquifex aeolicus in complex with the Fab fragment 3wkn 
3WKN 3wlw 
3WLW 3wn5 
3WN5 3wo3 
3WO3 3wo4 
3WO4 3wsq 
3WSQ 3wuw 
3WUW 3x2d 
3X2D 3x3f 
3X3F 3x3g 
3X3G 3zdx 
Integrin alphaIIB beta3 headpiece and RGD peptide complex 3zdy 
Integrin alphaIIB beta3 headpiece and RGD peptide complex 3zdz 
Integrin alphaIIB beta3 headpiece and RGD peptide complex 3ze0 
Integrin alphaIIB beta3 headpiece and RGD peptide complex 3ze1 
Integrin alphaIIB beta3 headpiece and RGD peptide complex 3ze2 
Integrin alphaIIB beta3 headpiece and RGD peptide complex 3zkm 
BACE2 FAB COMPLEX 3zkn 
BACE2 FAB INHIBITOR COMPLEX 3zl4 
Antibody structural organization: Role of kappa - lambda chain constant domain switch in catalytic functionality 3zo0 
Mouse IgG2a in complex with mouse TRIM21 PRYSPRY 3ztj 
Structure of influenza A neutralizing antibody selected from cultures of single human plasma cells in complex with human H3 Influenza haemagglutinin. 3ztn 
Structure of influenza A neutralizing antibody selected from cultures of single human plasma cells in complex with human H1 Influenza haemagglutinin. 4a6y 
CRYSTAL STRUCTURE OF FAB FRAGMENT OF ANTI-(4-HYDROXY-3-NITROPHENYL) -ACETYL MURINE GERMLINE ANTIBODY BBE6.12H3 4acp 
Deactivation of human IgG1 Fc by endoglycosidase treatment 4adf 
CRYSTAL STRUCTURE OF THE HUMAN COLONY-STIMULATING FACTOR 1 (hCSF-1) CYTOKINE IN COMPLEX WITH THE VIRAL RECEPTOR BARF1 4adq 
CRYSTAL STRUCTURE OF THE MOUSE COLONY-STIMULATING FACTOR 1 (MCSF-1) CYTOKINE IN COMPLEX WITH THE VIRAL RECEPTOR BARF1 4aeh 
Crystal structure of the anti-AaHI Fab9C2 antibody 4aei 
Crystal structure of the AaHII-Fab4C1 complex 4ag4 
Crystal structure of a DDR1-Fab complex 4al8 
Structure of Dengue virus DIII in complex with Fab 2H12 4ala 
Structure of Dengue virus DIII in complex with Fab 2H12 4am0 
Structure of Dengue virus strain 4 DIII in complex with Fab 2H12 4amk 
Fab fragment of antiporphrin antibody 13g10 4apq 
Crystal structure of autoreactive-Valpha14-Vbeta6 NKT TCR in complex with CD1d-sulfatide 4at6 
Fab fragment of antiporphyrin antibody 14H7 4b7i 
Crystal Structure of Human IgG Fc Bearing Hybrid-type Glycans 4ba8 
High resolution NMR structure of the C mu3 domain from IgM 4bfi 
Structure of the complex of the extracellular portions of mouse CD200R and mouse CD200 4bh7 
CRYSTAL STRUCTURE OF GERMLINE ANTIBODY 36-65 IN COMPLEX WITH PEPTIDE PPYPAWHAPGNI 4bh8 
CRYSTAL STRUCTURE OF GERMLINE ANTIBODY 36-65 IN COMPLEX WITH PEPTIDE GDPRPSYISHLL 4bjl 
LOCW, A LAMBDA 1 TYPE LIGHT-CHAIN DIMER (BENCE-JONES PROTEIN) CRYSTALLIZED IN DISTILLED WATER 4bkl 
Crystal structure of the arthritogenic antibody M2139 (Fab fragment) in complex with the triple-helical J1 peptide 4bm7 
Crystal Structure of IgG Fc F241A mutant with native glycosylation 4bsv 
Heterodimeric Fc Antibody Azymetric Variant 1 4bsw 
Heterodimeric Fc Antibody Azymetric Variant 2 4byh 
Crystal structure of sialylated IgG Fc 4bz1 
4BZ1 4bz2 
4BZ2 4c2i 
Cryo-EM structure of Dengue virus serotype 1 complexed with Fab fragments of human antibody 1F4 4c54 
Crystal structure of recombinant human IgG4 Fc 4c55 
Crystal structure of serum-derived human IgG4 Fc 4c56 
4C56 4c83 
Crystal Structure of the IgG2a LPT3 in complex with an 8-sugar inner core analogue of Neisseria meningitidis 4cad 
Mechanism of farnesylated CAAX protein processing by the integral membrane protease Rce1 4cbp 
4CBP 4cc8 
Pre-fusion structure of trimeric HIV-1 envelope glycoprotein determined by cryo-electron microscopy 4cdh 
4CDH 4cmh 
4CMH 4cmm 
Structure of human CD47 in complex with human Signal Regulatory Protein (SIRP) alpha v1 4cni 
Crystal structure of the Fab portion of Olokizumab in complex with IL- 6 4d2n 
4D2N 4d3c 
4D3C 4d9l 
Fab structure of anti-HIV-1 gp120 V2 mAb 697 4d9q 
Inhibiting Alternative Pathway Complement Activation by Targeting the Exosite on Factor D 4d9r 
Inhibiting Alternative Pathway Complement Activation by Targeting the Exosite on Factor D 4dag 
Structure of the Human Metapneumovirus Fusion Protein with Neutralizing Antibody Identifies a Pneumovirus Antigenic Site 4dcq 
Crystal Structure of the Fab Fragment of 3B5H10, an Antibody-Specific for Extended Polyglutamine Repeats (orthorhombic form) 4dep 
Structure of the IL-1b signaling complex 4dgi 
Structure of POM1 FAB fragment complexed with human PrPc Fragment 120-230 4dgv 
Structure of the Hepatitis C virus envelope glycoprotein E2 antigenic region 412-423 bound to the broadly neutralizing antibody HCV1, P2(1) form 4dgy 
Structure of the Hepatitis C virus envelope glycoprotein E2 antigenic region 412-423 bound to the broadly neutralizing antibody HCV1, C2 form 4dkd 
Crystal Structure of Human Interleukin-34 Bound to Human CSF-1R 4dke 
Crystal Structure of Human Interleukin-34 Bound to FAb1.1 4dkf 
Crystal Structure of Human Interleukin-34 Bound to FAb2 4dn3 
Crystal structure of anti-mcp-1 antibody cnto888 4dn4 
Crystal structure of the complex between cnto888 fab and mcp-1 mutant p8a 4dqo 
Crystal Structure of PG16 Fab in Complex with V1V2 Region from HIV-1 strain ZM109 4dtg 
Hemostatic effect of a monoclonal antibody mAb 2021 blocking the interaction between FXa and TFPI in a rabbit hemophilia model 4dvb 
The crystal structure of the Fab fragment of pro-uPA antibody mAb-112 4dvr 
Crystal structure of YU2 gp120 core in complex with Fab 48d and NBD-557 4dw2 
The crystal structure of uPA in complex with the Fab fragment of mAb-112 4dz8 
human IgG1 Fc fragment Heterodimer 4dzb 
Mucosal-associated invariant T cell receptor, Valpha7.2Jalpha33-Vbeta2 4e41 
Structural basis for the recognition of mutant self by a tumor-specific, MHC class II-restricted T cell receptor G4 4e42 
Structural basis for the recognition of mutant self by a tumor-specific, MHC class II-restricted T cell receptor G4 4ebq 
Fab structure of anti-Vaccinia virus D8L antigen mouse IgG2a LA5 4edw 
Nerve Growth Factor in Complex with Fab from humanized version of mouse mAb 911 (tanezumab) 4edx 
Nerve Growth Factor in Complex with Fab from mouse mAb 911 4ei5 
Crystal Structure of XV19 TCR in complex with CD1d-sulfatide C24:1 4ei6 
Structure of XV19 Valpha1-Vbeta16 Type-II Natural Killer T cell receptor 4elk 
Crystal structure of the Hy19.3 type II NKT TCR 4elm 
Crystal structure of the mouse CD1d-lysosulfatide-Hy19.3 TCR complex 4en3 
Crystal structure of a human Valpha24(-) NKT TCR in complex with CD1d/alpha-galactosylceramide 4ene 
Structure of the N- and C-terminal trimmed ClC-ec1 Cl-/H+ antiporter and Fab Complex 4eow 
Crystal structure of a disease-associated anti-human GM-CSF autoantibody MB007 4ers 
A Molecular Basis for Negative Regulation of the Glucagon Receptor 4etq 
Vaccinia virus D8L IMV envelope protein in complex with Fab of murine IgG2a LA5 4eup 
The complex between TCR JKF6 and human Class I MHC HLA-A2 presenting the MART-1(27-35)(A27L) peptide 4evn 
Crystal Structure of Fab CR6261 (somatic heavy chain with germline-reverted light chain) 4exp 
Structure of mouse Interleukin-34 in complex with mouse FMS 4f15 
Molecular basis of infectivity of 2009 pandemic H1N1 influenza A viruses 4f2m 
Crystal structure of a TGEV coronavirus Spike fragment in complex with the TGEV neutralizing monoclonal antibody 1AF10 4f33 
Crystal Structure of therapeutic antibody MORAb-009 4f37 
Structure of the tethered N-terminus of Alzheimer's disease A peptide 4f3f 
Crystal Structure of Msln7-64 MORAb-009 FAB complex 4f57 
Fab structure of a neutralizing antibody L1 from an early subtype A HIV-1 infected patient 4f58 
Fab structure of a neutralizing antibody L3 from an early subtype A HIV-1 infected patient 4fa8 
Multi-pronged modulation of cytokine signaling 4fab 
THREE-DIMENSIONAL STRUCTURE OF A FLUORESCEIN-FAB COMPLEX CRYSTALLIZED IN 2-METHYL-2,4-PENTANEDIOL 4ffv 
Crystal Structure of Dipeptidyl Peptidase IV (DPP4, DPP-IV, CD26) in Complex with 11A19 Fab 4ffw 
Crystal Structure of Dipeptidyl Peptidase IV (DPP4, DPP-IV, CD26) in Complex with Fab + sitagliptin 4ffz 
Crystal Structure of DENV1-E111 fab fragment bound to DENV-1 DIII (Western Pacific-74 strain). 4fg6 
Structure of EcCLC E148A mutant in Glutamate 4fmf 
Crystal structure of human nectin-1 full ectodomain (D1-D3) 4fmk 
Crystal structure of mouse nectin-2 extracellular fragment D1-D2 4fn0 
Crystal structure of mouse nectin-2 extracellular fragment D1-D2, 2nd crystal form 4fnl 
Crystal structure of broadly neutralizing antibody C05 4fom 
Crystal structure of human nectin-3 full ectodomain (D1-D3) 4fp8 
Crystal structure of broadly neutralizing antibody C05 bound to H3 influenza hemagglutinin, HA1 subunit 4fq1 
Crystal Structure of PGT121 Fab 4fq2 
Crystal Structure of 10-1074 Fab 4fqc 
Crystal Structure of PGT121 Fab Bound to a complex-type sialylated N-glycan 4fqh 
Crystal Structure of Fab CR9114 4fqi 
Crystal Structure of Fab CR9114 in Complex with a H5N1 influenza virus hemagglutinin 4fqj 
Influenza B/Florida/4/2006 hemagglutinin Fab CR8071 complex 4fqk 
Influenza B/Brisbane/60/2008 hemagglutinin Fab CR8059 complex 4fql 
Influenza B HA Antibody (Fab) CR8033 4fqp 
Crystal structure of human Nectin-like 5 full ectodomain (D1-D3) 4fqq 
Crystal Structure of Germline Antibody PGT121-GL Fab 4fqr 
Crystal structure of broadly neutralizing antibody C05 bound to H3 influenza hemagglutinin 4fqv 
Crystal structure of broadly neutralizing antibody CR9114 bound to H7 influenza hemagglutinin 4fqy 
Crystal structure of broadly neutralizing antibody CR9114 bound to H3 influenza hemagglutinin 4fs0 
Crystal structure of mutant F136D of mouse nectin-2 extracellular fragment D1-D2 4ftv 
The complex between the high affinity version of A6 TCR (A6c134) and human Class I MHC HLA-A2 with the bound TAX nonameric peptide 4fz8 
Crystal structure of C11 Fab, an ADCC mediating anti-HIV-1 antibody. 4fze 
Crystal structure of N26_i1 Fab, an ADCC mediating anti-HIV-1 antibody. 4g3y 
Crystal structure of TNF-alpha in complex with Infliximab Fab fragment 4g5z 
Crystal structure of the therapeutical antibody fragment of canakinumab in its unbound state 4g6a 
Structure of the Hepatitis C virus envelope glycoprotein E2 antigenic region 412-423 bound to the broadly neutralizing antibody AP33 4g6f 
Crystal Structure of 10E8 Fab in Complex with an HIV-1 gp41 Peptide 4g6j 
Crystal structure of human IL-1beta in complex with the therapeutic antibody binding fragment of canakinumab 4g6k 
Crystal structure of the therapeutic antibody binding fragment of gevokizumab in its unbound state 4g6m 
Crystal strucure of human IL-1beta in complex with therapeutic antibody binding fragment of gevokizumab 4g7v 
Crystal structure of voltage sensing domain of Ci-VSP with fragment antibody (R217E, 2.5 A) 4g7y 
Crystal structure of voltage sensing domain of Ci-VSP with fragment antibody (R217E, 2.8 A) 4g80 
Crystal structure of voltage sensing domain of Ci-VSP with fragment antibody (WT, 3.8 A) 4g8e 
Crystal Structure of clone18 TCR 4g8f 
Crystal Structure of clone42 TCR 4g8g 
Crystal Structure of C12C TCR-HA B2705-KK10 4g9f 
Crystal Structure of C12C TCR-HLAB2705-KK10-L6M 4gaf 
Crystal structure of EBI-005, a chimera of human IL-1beta and IL-1Ra, bound to human Interleukin-1 receptor type 1 4gag 
Structure of the broadly neutralizing antibody AP33 in complex with its HCV epitope (E2 residues 412-423) 4gaj 
Structure of the broadly neutralizing antibody AP33 in complex with its HCV epitope (E2 residues 411-424) 4gay 
Structure of the broadly neutralizing antibody AP33 4gg6 
Protein complex 4gg8 
Immune Receptor 4gkz 
HA1.7, a MHC class II restricted TCR specific for haemagglutinin 4glr 
Structure of the anti-ptau Fab (pT231/pS235_1) in complex with phosphoepitope pT231/pS235 4gms 
Crystal structure of heterosubtypic Fab S139/1 in complex with influenza A H3 hemagglutinin 4gmt 
Crystal structure of heterosubtypic Fab S139/1 4gq9 
Chikungunya virus neutralizing antibody 9.8B Fab fragment 4grl 
Crystal structure of a autoimmune TCR-MHC complex 4grm 
The crystal structure of the high affinity TCR A6 4gsd 
H5.3 Fab Structure 4gw1 
cQFD Meditope 4gw4 
Crystal structure of 3BNC60 Fab with P61A mutation 4gw5 
cQYN meditope - Cetuximab Fab 4gxu 
Crystal structure of antibody 1F1 bound to the 1918 influenza hemagglutinin 4gxv 
Crystal structure of anti-influenza virus antibody 1F1 4h20 
Crystal Structure and Computational Modeling of the Fab Fragment from the Protective anti-Ricin Monoclonal Antibody RAC18 4h88 
Structure of POM1 FAB fragment complexed with mouse PrPc Fragment 120-230 4h8w 
Crystal structure of non-neutralizing and ADCC-potent antibody N5-i5 in complex with HIV-1 clade A/E gp120 and sCD4. 4haf 
Crystal structure of fc-fragment of human IgG2 antibody (primitive crystal form) 4hag 
Crystal structure of fc-fragment of human IgG2 antibody (centered crystal form) 4hbc 
Crystal structure of a conformation-dependent rabbit IgG Fab specific for amyloid prefibrillar oligomers 4hc1 
Crystal structure of a loop deleted mutant of human MAdCAM-1 D1D2 complexed with Fab 10G3 4hcr 
Crystal structure of human MAdCAM-1 D1D2 complexed with Fab PF-547659 4hdi 
Crystal Structure of 3E5 IgG3 FAB from mus musculus 4hf5 
Crystal structure of Fab 8F8 in complex a H2N2 influenza virus hemagglutinin 4hfu 
Crystal structure of Fab 8M2 in complex with a H2N2 influenza virus hemagglutinin 4hfw 
Anti Rotavirus Antibody 4hg4 
Crystal structure of Fab 2G1 in complex with a H2N2 influenza virus hemagglutinin 4hgw 
Crystal structure of S25-2 in complex with a 5,6-dehydro-Kdo disaccharide 4hh8 
Crystal structure of bovine butyrophilin 4hh9 
Anti-Human Cytomegalovirus (HCMV) Fab KE5 4hha 
Anti-Human Cytomegalovirus (HCMV) Fab KE5 with epitope peptide AD-2S1 4hie 
Anti-Streptococcus pneumoniae 23F Fab 023.102 4hih 
Anti-Streptococcus pneumoniae 23F Fab 023.102 with bound rhamnose. 4hii 
Anti-Streptococcus pneumoniae 23F Fab 023.102 with bound rhamnose-galactose 4hij 
Anti-Streptococcus pneumoniae 23F Fab 023.102 with bound L-rhamnose-(1-2)-alpha-D-galactose-(3-O)-phosphate-2-glycerol 4hix 
Crystal structure of a humanised 3D6 Fab bound to amyloid beta peptide 4hj0 
Crystal structure of the human GIPr ECD in complex with Gipg013 Fab at 3-A resolution 4hjg 
Meditope-enabled trastuzumab 4hjj 
Structure Reveals Function of the Dual Variable Domain Immunoglobulin (DVD-Ig) Molecule 4hk0 
UCA Fab (unbound) from CH65-CH67 Lineage 4hk3 
I2 Fab (unbound) from CH65-CH67 Lineage 4hkb 
CH67 Fab (unbound) from the CH65-67 Lineage 4hkx 
Influenza hemagglutinin in complex with CH67 Fab 4hkz 
Trastuzumab Fab complexed with Protein L and Protein A fragments 4hlz 
Crystal Structure of Fab C179 in Complex with a H2N2 influenza virus hemagglutinin 4hpo 
Crystal structure of RV144-elicited antibody CH58 in complex with V2 peptide 4hpy 
Crystal structure of RV144-elicited antibody CH59 in complex with V2 peptide 4hqq 
Crystal structure of RV144-elicited antibody CH58 4hs6 
Hepatitus C envelope glycoprotein E2 fragment 412-423 with humanized and affinity-matured antibody MRCT10.v362 4hs8 
Hepatitus C envelope glycoprotein E2 fragment 412-423 with humanized and affinity-matured antibody hu5B3.v3 4ht1 
Human TWEAK in complex with the Fab fragment of a neutralizing antibody 4hwb 
Crystal structure of ectodomain 3 of the IL-13 receptor alpha 1 in complex with a human neutralizing monoclonal antibody fragment 4hwe 
Crystal structure of ectodomain 3 of the IL-13 receptor alpha1 in complex with a human neutralizing monoclonal antibody fragment 4hxa 
Crystal structure of 13D9 FAB 4hxb 
Crystal structure of 6B9 FAB 4hzl 
Neutralizing antibody mAb#8 in complex with the Epitope II of HCV E2 envelope protein 4i0k 
Crystal structure of murine B7-H3 extracellular domain 4i18 
Crystal structure of human prolactin receptor complexed with Fab fragment 4i2x 
Crystal structure of Signal Regulatory Protein gamma (SIRP-gamma) in complex with FabOX117 4i3r 
Crystal structure of the outer domain of HIV-1 gp120 in complex with VRC-PG04 space group P3221 4i3s 
Crystal structure of the outer domain of HIV-1 gp120 in complex with VRC-PG04 space group P21 4i77 
Lebrikizumab Fab bound to IL-13 4i9w 
Human two pore domain K+ channel TRAAK (K2P4.1) - Fab complex structure 4idj 
S.Aureus a-hemolysin monomer in complex with Fab 4iiq 
Crystal structure of a human MAIT TCR in complex with bovine MR1 4ij3 
Oxidoreductase Fragment of Human QSOX1 in Complex with a FAB Fragment from an Anti- Human QSOX1 Antibody 4imk 
Uncrossed Fab binding to human Angiopoietin 2 4iml 
CrossFab binding to human Angiopoietin 2 4iof 
Crystal structure analysis of Fab-bound human Insulin Degrading Enzyme (IDE) 4ioi 
Meditope-enabled trastuzumab in complex with CQFDLSTRRLKC 4irj 
Structure of the mouse CD1d-4ClPhC-alpha-GalCer-iNKT TCR complex 4irs 
Structure of the mouse CD1d-PyrC-alpha-GalCer-iNKT TCR complex 4irz 
Crystal structure of A4b7 headpiece complexed with Fab Natalizumab 4isv 
Crystal structure of the Fab FRAGMENT OF 1C2, A MONOCLONAL ANTIBODY SPECIFIC FOR POLY-GLUTAMINE 4j12 
monomeric Fc 4j1u 
Crystal structure of antibody 93F3 unstable variant 4j4p 
The complex of human IgE-Fc with two bound Fab fragments 4j6r 
Crystal structure of broadly and potently neutralizing antibody VRC23 in complex with HIV-1 gp120 4j8r 
Structure of an octapeptide repeat of the prion protein bound to the POM2 Fab antibody fragment 4jam 
Crystal structure of broadly neutralizing anti-hiv-1 antibody ch103 4jan 
crystal structure of broadly neutralizing antibody CH103 in complex with HIV-1 gp120 4jb9 
Crystal structure of antibody VRC06 in complex with HIV-1 gp120 core 4jdt 
Crystal structure of chimeric germ-line precursor of NIH45-46 Fab in complex with gp120 of 93TH057 HIV-1 4jdv 
Crystal structure of germ-line precursor of NIH45-46 Fab 4jfd 
Preservation of peptide specificity during TCR-MHC contact dominated affinity enhancement of a melanoma-specific TCR 4jfe 
Preservation of peptide specificity during TCR-MHC contact dominated affinity enhancement of a melanoma-specific TCR 4jff 
Preservation of peptide specificity during TCR-MHC contact dominated affinity enhancement of a melanoma-specific TCR 4jfh 
High Affinity alpha24-beta17 T Cell Receptor for A2 HLA-Melanoma peptide complex 4jfx 
Structure of phosphotyrosine (pTyr) scaffold bound to pTyr peptide 4jfy 
Apo structure of phosphotyrosine (pYAb) scaffold 4jfz 
Structure of phosphoserine (pSAb) scaffold bound to pSer peptide 4jg0 
Structure of phosphoserine/threonine (pSTAb) scaffold bound to pSer peptide 4jg1 
Structure of phosphoserine/threonine (pSTAb) scaffold bound to pThr peptide 4jha 
Crystal Structure of RSV-Neutralizing Human Antibody D25 4jhw 
Crystal Structure of Respiratory Syncytial Virus Fusion Glycoprotein Stabilized in the Prefusion Conformation by Human Antibody D25 4jj5 
CRYSTAL STRUCTURE OF THE Fab FRAGMENT OF 1C2, A MONOCLONAL ANTIBODY SPECIFIC for POLY-GLUTAMINE 4jkp 
Restricting HIV-1 Pathways for Escape using Rationally-Designed Anti-HIV-1 Antibodies 4jlr 
Crystal structure of a designed Respiratory Syncytial Virus Immunogen in complex with Motavizumab 4jm2 
Crystal Structure of PGT 135 Fab in Complex with gp120 Core Protein from HIV-1 Strain JR-FL Bound to CD4 and 17b Fab 4jm4 
Crystal Structure of PGT 135 Fab 4jn1 
An Antidote for Dabigatran 4jn2 
An Antidote for Dabigatran 4jo1 
Crystal structure of rabbit mAb R56 Fab in complex with V3 crown of HIV-1 JR-FL gp120 4jo2 
Crystal structure of rabbit mAb R56 Fab in complex with V3 crown of HIV-1 Consensus A gp120 4jo3 
Crystal structure of rabbit mAb R20 Fab in complex with V3 C-terminus of HIV-1 Consensus B gp120 4jo4 
Crystal structure of rabbit mAb R20 Fab 4jpi 
Crystal structure of a putative VRC01 germline precursor Fab 4jpk 
Crystal structure of the germline-targeting HIV-1 gp120 engineered outer domain eOD-GT6 in complex with a putative VRC01 germline precursor Fab 4jpv 
Crystal structure of broadly and potently neutralizing antibody 3bnc117 in complex with hiv-1 gp120 4jpw 
Crystal structure of broadly and potently neutralizing antibody 12a21 in complex with hiv-1 strain 93th057 gp120 mutant 4jqi 
Structure of active beta-arrestin1 bound to a G protein-coupled receptor phosphopeptide 4jr9 
Crystal structure of nitrate/nitrite exchanger NarK 4jre 
Crystal structure of nitrate/nitrite exchanger NarK with nitrite bound 4jrx 
Crystal Structure of CA5 TCR-HLA B*3505-LPEP complex 4jry 
Crystal Structure of SB47 TCR-HLA B*3505-LPEP complex 4jy4 
Crystal structure of human Fab PGT121, a broadly reactive and potent HIV-1 neutralizing antibody 4jy5 
Crystal structure of human Fab PGT122, a broadly reactive and potent HIV-1 neutralizing antibody 4jy6 
Crystal structure of human Fab PGT123, a broadly reactive and potent HIV-1 neutralizing antibody 4jzj 
Crystal Structure of Receptor-Fab Complex 4jzn 
Three dimensional structure of broadly neutralizing human anti - Hepatitis C virus (HCV) glycoprotein E2 Fab fragment HC84-1 4jzo 
Three dimensional structure of broadly neutralizing human anti - Hepatitis C virus (HCV) glycoprotein E2 Fab fragment HC84-27 4k0v 
Structural basis for angiopoietin-1 mediated signaling initiation 4k23 
Structure of anti-uPAR Fab ATN-658 4k24 
Structure of anti-uPAR Fab ATN-658 in complex with uPAR 4k2u 
Crystal structure of PfEBA-175 F1 in complex with R218 antibody Fab fragment 4k3d 
Crystal structure of bovine antibody BLV1H12 with ultralong CDR H3 4k3e 
Crystal structure of bovine antibody BLV5B8 with ultralong CDR H3 4k3j 
Crystal structure of Onartuzumab Fab in complex with MET and HGF-beta 4k7p 
Generation and Characterization of a Unique Reagent that Recognizes a Panel of Recombinant Human Monoclonal Antibody Therapeutics in the Presence of Endogenous Human IgG 4k8r 
An Antibody Against the C-terminal Domain of PCSK9 lowers LDL Cholesterol Levels in vivo 4k94 
Crystal structure of KIT D4D5 fragment in complex with anti-Kit antibody Fab19 4k9e 
Crystal structure of KIT D4D5 fragment in complex with anti-Kit antibodies Fab79D 4kaq 
4KAQ 4kc3 
Cytokine/receptor binary complex 4kht 
Triple helix bundle of GP41 complexed with fab 8066 4khx 
Crystal structure of gp41 helix complexed with antibody 8062 4ki5 
Cystal structure of human factor VIII C2 domain in a ternary complex with murine inhbitory antibodies 3E6 and G99 4kjp 
Structure of the CLC-ec1 deltaNC construct in the absence of halide 4kjq 
Structure of the CLC-ec1 deltaNC construct in 100mM fluoride 4kjw 
Structure of the CLC-ec1 deltaNC construct in 100mM fluoride and 20mM bromide 4kjy 
Complex of high-affinity SIRP alpha variant FD6 with CD47 4kk5 
Structure of the CLC-ec1 deltaNC construct in 20mM fluoride and 20mM bromide 4kk6 
Structure of CLC-ec1 deltaNC construct in 20mM Bromide 4kk8 
Structure of the E148Q mutant of CLC-ec1 deltaNC construct in 100mM fluoride 4kk9 
Structure of the E148A mutant of CLC-ec1 deltaNC construct in 100mM fluoride and 2mM Bromide 4kka 
Structure of the E148A mutant of CLC-ec1 deltaNC construct in 100mM fluoride and 20mM Bromide 4kkb 
Structure of the E148A mutant of CLC-ec1 deltaNC construct in 20mM fluoride and 20mM Bromide 4kkc 
Structure of the E148A mutant of CLC-ec1 deltaNC construct in 20mM Bromide 4kkl 
Structure of the E148A mutant of CLC-ec1 delta NC construct in 100mM fluoride 4kmt 
Crystal structure of human germline antibody 5-51/O12 4kph 
Structure of the Fab fragment of N62, a protective monoclonal antibody to the nonreducing end of Francisella tularensis O-antigen 4kq3 
Crystal structure of Anti-IL-17A antibody CNTO3186 4kq4 
Crystal structure of Anti-IL-17A antibody CNTO7357 4kro 
Nanobody/VHH domain EgA1 in complex with the extracellular region of EGFR 4krp 
Nanobody/VHH domain 9G8 in complex with the extracellular region of EGFR 4ktd 
Fab fragment of HIV vaccine-elicited CD4bs-directed antibody, GE136, from non-human primate 4kte 
Fab fragment of HIV vaccine-elicited CD4bs-directed antibody, GE148, from non-human primate 4ku1 
Role of the hinge and C-gamma-2/C-gamma-3 interface in immunoglobin G1 Fc domain motions: implications for Fc engineering 4kuc 
Crystal structure of ricin-A chain in complex with the antibody 6C2 4kuz 
Crystal structure of anti-emmprin antibody 4A5 Fab in trigonal form 4kvc 
2H2 Fab fragment of immature Dengue virus 4kvn 
Crystal structure of Fab 39.29 in complex with Influenza Hemagglutinin A/Perth/16/2009 (H3N2) 4kxz 
4KXZ 4ky1 
humanized HP1/2 Fab 4kzd 
4KZD 4kze 
4KZE 4l3e 
4L3E 4l4j 
Crystal structure of fc-fragment of human IgG2-Sigma antibody 4l4t 
Structure of human MAIT TCR in complex with human MR1-6-FP 4l4v 
Structure of human MAIT TCR in complex with human MR1-RL-6-Me-7-OH 4l5f 
Crystal Structure of DENV1-E106 Fab bound to DENV-1 Envelope protein DIII 4l8s 
Crystal structure of a human Valpha7.2/Vbeta13.3 MAIT TCR in complex with bovine MR1 4l9l 
Crystal structure of a human Valpha7.2/Vbeta13.2 MAIT TCR in complex with bovine MR1 4lbe 
Structure of KcsA with R122A mutation 4lcc 
Crystal structure of a human MAIT TCR in complex with a bacterial antigen bound to humanized bovine MR1 4lcu 
Structure of KcsA with E118A mutation 4lcw 
The structure of human MAIT TCR in complex with MR1-K43A-RL-6-Me-7OH 4leo 
Crystal structure of anti-HER3 Fab RG7116 in complex with the extracellular domains of human Her3 (ERBB3) 4lex 
Unliganded crystal structure of mAb7 4lf3 
Inhibitory Mechanism of an Allosteric Antibody Targeting the Glucagon Receptor 4lfh 
Crystal Structure of 9C2 TCR 4lhu 
Crystal Structure of 9C2 TCR bound to CD1d 4liq 
4LIQ 4lkc 
An Antibody Against the C-terminal Domain of PCSK9 lowers LDL Cholesterol Levels in vivo 4lkx 
Humanized antibody 4B12 Fab complexed with a CemX segment 4llu 
Structure of Pertuzumab Fab with light chain Clambda at 2.16A 4llw 
Crystal structure of Pertuzumab Clambda Fab with variable domain redesign (VRD2) at 1.95A 4lly 
Crystal structure of Pertuzumab Clambda Fab with variable and constant domain redesigns (VRD2 and CRD2) at 1.6A 4lmq 
Development and Preclinical Characterization of a Humanized Antibody Targeting CXCL12 4lou 
Structure of the E148Q mutant of CLC-ec1 deltaNC construct in the absence of halide 4lp5 
Crystal structure of the full-length human RAGE extracellular domain (VC1C2 fragment) 4lqf 
4LQF 4lri 
Anti CMV Fab Fragment 4lsp 
Crystal structure of broadly and potently neutralizing antibody VRC-CH31 in complex with HIV-1 clade A/E gp120 93TH057 4lsq 
Crystal structure of broadly and potently neutralizing antibody VRC-CH31 in complex with HIV-1 clade A/E gp120 93TH057 with LOOP D and Loop V5 from clade A strain 3415_v1_c1 4lsr 
Crystal structure of broadly and potently neutralizing antibody VRC-CH31 in complex with HIV-1 clade A/E stran 93TH057 gp120 with LOOP D and Loop V5 from clade A strain KER_2018_11 4lss 
Crystal structure of broadly and potently neutralizing antibody VRC01 in complex with HIV-1 clade A strain KER_2018_11 gp120 4lst 
Crystal structure of broadly and potently neutralizing antibody VRC01 in complex with HIV-1 clade C strain ZM176.66 gp120 4lsu 
Crystal structure of broadly and potently neutralizing antibody VRC-PG20 in complex with HIV-1 clade A/E 93TH057 gp120 4lsv 
Crystal structure of broadly and potently neutralizing antibody 3BNC117 in complex with HIV-1 clade C C1086 gp120 4lu5 
4LU5 4lvh 
Insight into highly conserved H1 subtype-specific epitopes in influenza virus hemagglutinin 4lvn 
Crystal structure of PfSUB1-prodomain-NIMP.M7 Fab complex 4lvo 
Crystal structure of PfSUB1-prodomain-NIMP.M7 Fab complex with added CaCl2 4m1c 
4M1C 4m1d 
Crystal structure of anti-HIV-1 Fab 447-52D in complex with V3 cyclic peptide MN 4m1g 
4M1G 4m43 
Crystal structure of anti-rabies glycoprotein Fab 523-11 4m48 
X-ray structure of dopamine transporter elucidates antidepressant mechanism 4m5y 
Crystal structure of broadly neutralizing Fab 5J8 4m5z 
Crystal structure of broadly neutralizing antibody 5J8 bound to 2009 pandemic influenza hemagglutinin, HA1 subunit 4m61 
Crystal structure of unliganded anti-DNA Fab A52 4m6m 
Crystal structure of anti-IL-23 antibody CNTO1959 at pH 9.5 4m6n 
4M6N 4m6o 
Crystal structure of anti-NGF antibody CNTO7309 4m7j 
Crystal structure of S25-26 in complex with Kdo(2.8)Kdo(2.4)Kdo trisaccharide 4m7k 
Crystal structure of anti-tissue factor antibody 10H10 4m7l 
4M7L 4m7z 
Unliganded 1 crystal structure of S25-26 Fab 4m93 
Unliganded 2 crystal structure of S25-26 Fab 4ma1 
Unliganded 3 crystal structure of S25-26 Fab 4ma3 
Crystal structure of anti-hinge rabbit antibody C2095 4ma7 
Crystal structure of mouse prion protein complexed with Promazine 4ma8 
Crystal structure of mouse prion protein complexed with Chlorpromazine 4mau 
Crystal structure of anti-ST2L antibody C2244 4may 
Crystal structure of an immune complex 4mhh 
Crystal structure of Fab H5M9 in complex with influenza virus hemagglutinin from A/Viet Nam/1203/2004 (H5N1) 4mhj 
Crystal structure of Fab H5M9 in complex with influenza virus hemagglutinin from A/goose/Guangdong/1/96 (H5N1) 4mji 
4MJI 4mng 
Structure of the DP10.7 TCR with CD1d-sulfatide 4mnh 
Structure of the DP10.7 TCR 4mnq 
TCR-peptide specificity overrides affinity enhancing TCR-MHC interactions 4mqx 
CLC-ec1 Fab Complex Cysless A399C-A432C mutant 4ms8 
4MS8 4msw 
Y78 ester mutant of KcsA in high K+ 4mvb 
4MVB 4mwf 
Structure of Hepatitis C Virus Envelope Glycoprotein E2 core bound to broadly neutralizing antibody AR3C 4mxq 
4MXQ 4mxv 
Structure of Lymphotoxin alpha bound to anti-LTa Fab 4mxw 
Structure of heterotrimeric lymphotoxin LTa1b2 bound to lymphotoxin beta receptor LTbR and anti-LTa Fab 4myw 
4MYW 4n0c 
4N0C 4n0u 
Ternary complex between Neonatal Fc receptor, serum albumin and Fc 4n0y 
4N0Y 4n5e 
4N5E 4n8c 
4N8C 4n8v 
Crystal structure of killer cell immunoglobulin-like receptor KIR2DS2 in complex with HLA-A 4n90 
4N90 4n9g 
Crystal Structure of a Computationally Designed RSV-Presenting Epitope Scaffold And Its Elicited Antibody 17HD9 4ncc 
Neutralizing antibody to murine norovirus 4nco 
Crystal Structure of the BG505 SOSIP gp140 HIV-1 Env trimer in Complex with the Broadly Neutralizing Fab PGT122 4ndm 
Structure of the AB18.1 TCR 4ngh 
4NGH 4nhc 
4NHC 4nhg 
Crystal Structure of 2G12 IgG Dimer 4nhh 
Structure of 2G12 IgG Dimer 4nhu 
4NHU 4nj9 
4NJ9 4nja 
4NJA 4nki 
4NKI 4nm4 
Crystal structure of broadly neutralizing antibody CR8043 4nm8 
Crystal structure of broadly neutralizing antibody CR8043 bound to H3 influenza hemagglutinin 4nnp 
4NNP 4no0 
4NO0 4np4 
Clostridium difficile toxin B CROP domain in complex with FAB domains of neutralizing antibody bezlotoxumab 4npy 
Crystal structure of germline Fab PGT121, a putative precursor of the broadly reactive and potent HIV-1 neutralizing antibody 4nqc 
Crystal structure of TCR-MHC ternary complex and covalently bound 5-(2-oxopropylideneamino)-6-D-ribitylaminouracil 4nqd 
Crystal structure of TCR-MHC ternary complex and non-covalently bound 5-(2-oxopropylideneamino)-6-D-ribitylaminouracil 4nqe 
Crystal structure of TCR-MHC ternary complex bound to 5-(2-oxoethylideneamino)-6-D-ribitylaminouracil 4nqs 
Knob-into-hole IgG Fc 4nqt 
anti-parallel Fc-hole(T366S/L368A/Y407V) homodimer 4nqu 
anti-parallel Fc-knob (T366W) homodimer 4nrx 
Crystal Structure of HIV-1 Neutralizing Antibody m66 in complex with gp41 MPER peptide 4nry 
Crystal Structure of HIV-1 Neutralizing Antibody m66 4nrz 
Crystal Structure of HIV-1 Neutralizing Antibody m66.6 4nug 
Crystal structure of HIV-1 broadly neutralizing antibody PGT151 4nuj 
Crystal structure of HIV-1 broadly neutralizing antibody PGT152 4nwt 
4NWT 4nwu 
4NWU 4nyl 
4NYL 4nzr 
Crystal structure of the antibody-binding region of Protein M (Protein M TD) in complex with anti-HIV antibody PGT135 Fab 4nzt 
Crystal structure of the antibody-binding region of Protein M (Protein M TD) in complex with anti-infleunza hemagglutinin antibody CR9114 Fab 4nzu 
Crystal structure of the primary monoclonal antibody 13PL Fab' from a multiple myeloma patient 4o02 
AlphaVBeta3 integrin in complex with monoclonal antibody FAB fragment. 4o4y 
Crystal structure of the anti-hinge rabbit antibody 2095-2 in complex with IDES hinge peptide 4o51 
Crystal structure of the QAA variant of anti-hinge rabbit antibody 2095-2 in complex with IDES hinge peptide 4o58 
Crystal structure of broadly neutralizing antibody F045-092 in complex with A/Victoria/3/1975 (H3N2) influenza hemagglutinin 4o5i 
Crystal structure of broadly neutralizing antibody F045-092 in complex with A/Victoria/361/2011 (H3N2) influenza hemagglutinin 4o5l 
Crystal structure of broadly neutralizing antibody F045-092 4o9h 
4O9H 4oaw 
Fab structure of anti-HIV gp120 V2 mAb 2158 4ocr 
Crystal structure of human Fab CAP256-VRC26.01, a potent V1V2-directed HIV-1 neutralizing antibody 4ocs 
Crystal structure of human Fab CAP256-VRC26.10, a potent V1V2-directed HIV-1 neutralizing antibody 4ocw 
Crystal structure of human Fab CAP256-VRC26.06, a potent V1V2-directed HIV-1 neutralizing antibody 4ocx 
4OCX 4ocy 
4OCY 4od1 
Crystal structure of human Fab CAP256-VRC26.03, a potent V1V2-directed HIV-1 neutralizing antibody 4od2 
Crystal structure of the Fab fragment of an anti-DR5 antibody bound to DR5 4od3 
Crystal structure of human Fab CAP256-VRC26.07, a potent V1V2-directed HIV-1 neutralizing antibody 4odh 
Crystal structure of human Fab CAP256-VRC26.UCA, a potent V1V2-directed HIV-1 neutralizing antibody 4ods 
4ODS 4odt 
4ODT 4odu 
4ODU 4odv 
4ODV 4odw 
4ODW 4of8 
Crystal Structure of Rst D1-D2 4ofd 
Crystal Structure of mouse Neph1 D1-D2 4ofy 
Crystal Structure of the Complex of SYG-1 D1-D2 and SYG-2 D1-D4 4ogx 
4OGX 4ogy 
4OGY 4oi9 
4OI9 4oia 
4OIA 4oib 
4OIB 4oii 
West Nile Virus NS1 in complex with neutralizing 22NS1 antibody Fab 4ojf 
4OJF 4okv 
Crystal structure of anopheline anti-platelet protein with Fab antibody 4olu 
4OLU 4olv 
4OLV 4olw 
4OLW 4olx 
4OLX 4oly 
4OLY 4olz 
4OLZ 4om0 
4OM0 4om1 
4OM1 4onf 
4ONF 4ong 
4ONG 4onh 
4ONH 4oqt 
LINGO-1/Li81 Fab complex 4org 
Crystal structure of human Fab CAP256-VRC26.04, a potent V1V2-directed HIV-1 neutralizing antibody 4osu 
4OSU 4ot1 
4OT1 4otx 
4OTX 4ouu 
4OUU 4oz4 
4OZ4 4ozf 
JR5.1 protein complex 4ozg 
D2 protein complex 4ozh 
S16 protein complex 4ozi 
S2 protein complex 4p23 
4P23 4p2o 
Crystal structure of the 2B4 TCR in complex with 2A/I-Ek 4p2q 
Crystal structure of the 5cc7 TCR in complex with 5c2/I-Ek 4p2r 
Crystal structure of the 5cc7 TCR in complex with 5c1/I-Ek 4p2y 
4P2Y 4p3c 
4P3C 4p3d 
4P3D 4p46 
4P46 4p4k 
4P4K 4p59 
HER3 extracellular domain in complex with Fab fragment of MOR09825 4p5t 
4P5T 4p9h 
Crystal structure of 8ANC195 Fab in complex with gp120 of 93TH057 HIV-1 and soluble CD4 D1D2 4p9m 
Crystal structure of 8ANC195 Fab 4pb0 
4PB0 4pb9 
4PB9 4pgz 
4PGZ 4pj5 
4PJ5 4pj7 
4PJ7 4pj8 
4PJ8 4pj9 
4PJ9 4pja 
4PJA 4pjb 
4PJB 4pjc 
4PJC 4pjd 
4PJD 4pje 
4PJE 4pjf 
4PJF 4pjg 
4PJG 4pjh 
4PJH 4pji 
4PJI 4pjx 
4PJX 4plj 
4PLJ 4plk 
4PLK 4pou 
4POU 4poz 
4POZ 4pp1 
4PP1 4pp2 
4PP2 4prh 
Crystal structure of TK3 TCR-HLA-B*35:08-HPVG-D5 complex 4pri 
Crystal structure of TK3 TCR-HLA-B*35:08-HPVG complex 4prp 
Crystal structure of TK3 TCR-HLA-B*35:01-HPVG-Q5 complex 4ps4 
Crystal structure of the complex between IL-13 and M1295 FAB 4ptt 
4PTT 4ptu 
4PTU 4pub 
4PUB 4py7 
Crystal Structure of Fab 3.1 4py8 
Crystal structure of Fab 3.1 in complex with the 1918 influenza virus hemagglutinin 4q0x 
4Q0X 4q2z 
4Q2Z 4q5z 
4Q5Z 4q6i 
4Q6I 4q6y 
4Q6Y 4q74 
4Q74 4q7d 
4Q7D 4q9q 
4Q9Q 4q9r 
4Q9R 4qci 
4QCI 4qex 
4QEX 4qf1 
4QF1 4qgt 
4QGT 4qhk 
4QHK 4qhl 
4QHL 4qhm 
4QHM 4qhn 
4QHN 4qhu 
4QHU 4qnp 
4QNP 4qok 
4QOK 4qrp 
4QRP 4qrr 
4QRR 4qt5 
4QT5 4qth 
4QTH 4qti 
4QTI 4qww 
4QWW 4qxg 
4QXG 4qxu 
4QXU 4r0l 
4R0L 4r26 
4R26 4r2g 
4R2G 4r3z 
4R3Z 4r4b 
4R4B 4r4f 
4R4F 4r4h 
4R4H 4r4n 
4R4N 4r6u 
4R6U 4r7d 
4R7D 4r7n 
4R7N 4r8w 
4R8W 4r90 
4R90 4r96 
4R96 4r97 
4R97 4r9y 
4R9Y 4rau 
4RAU 4rbp 
4RBP 4rdq 
4RDQ 4rfe 
4RFE 4rfn 
4RFN 4rfo 
4RFO 4rgm 
4RGM 4rgn 
4RGN 4rgo 
4RGO 4rir 
4RIR 4ris 
4RIS 4rnr 
4RNR 4rqq 
4RQQ 4rqs 
4RQS 4rrp 
4RRP 4rwy 
4RWY 4rx4 
4RX4 4s1d 
4S1D 4s1q 
4S1Q 4s1r 
4S1R 4s1s 
4S1S 4s2s 
4S2S 4tnv 
4TNV 4tnw 
4TNW 4toy 
4TOY 4tpr 
4TPR 4tqe 
4TQE 4trp 
4TRP 4tsa 
4TSA 4tsb 
4TSB 4tsc 
4TSC 4ttd 
4TTD 4tuj 
4TUJ 4tuk 
4TUK 4tul 
4TUL 4tuo 
4TUO 4tvp 
4TVP 4u0r 
4U0R 4u1g 
4U1G 4u6g 
4U6G 4u6h 
4U6H 4u6v 
4U6V 4u7m 
4U7M 4uao 
4UAO 4ub0 
4UB0 4ubd 
4UBD 4udt 
4UDT 4udu 
4UDU 4uik 
4UIK 4uil 
4UIL 4uim 
4UIM 4uin 
4UIN 4uok 
4UOK 4uom 
4UOM 4ut6 
4UT6 4uta 
4UTA 4utb 
4UTB 4uuj 
4UUJ 4uv4 
4UV4 4uv7 
4UV7 4v10 
4V10 4w4n 
4W4N 4w4o 
4W4O 4wcy 
4WCY 4web 
4WEB 4wfe 
4WFE 4wff 
4WFF 4wfg 
4WFG 4wfh 
4WFH 4wht 
4WHT 4why 
4WHY 4wi2 
4WI2 4wi3 
4WI3 4wi4 
4WI4 4wi5 
4WI5 4wi6 
4WI6 4wi7 
4WI7 4wi8 
4WI8 4wi9 
4WI9 4wnq 
4WNQ 4wo4 
4WO4 4wrl 
4WRL 4wrm 
4WRM 4wuk 
4WUK 4wuu 
4WUU 4wv1 
4WV1 4ww1 
4WW1 4ww2 
4WW2 4wwi 
4WWI 4wwk 
4WWK 4wy7 
4WY7 4x0k 
4X0K 4x4m 
4X4M 4x6b 
4X6B 4x6c 
4X6C 4x6d 
4X6D 4x7s 
4X7S 4x7t 
4X7T 4x80 
4X80 4x83 
4X83 4x8j 
4X8J 4x8x 
4X8X 4x98 
4X98 4x99 
4X99 4x9b 
4X9B 4x9f 
4X9F 4x9g 
4X9G 4x9h 
4X9H 4x9i 
4X9I 4xak 
4XAK 4xaw 
4XAW 4xb7 
4XB7 4xb8 
4XB8 4xbe 
4XBE 4xbg 
4XBG 4xbp 
4XBP 4xc1 
4XC1 4xc3 
4XC3 4xcc 
4XCC 4xce 
4XCE 4xcf 
4XCF 4xcn 
4XCN 4xcy 
4XCY 4xgz 
4XGZ 4xh2 
4XH2 4xhj 
4XHJ 4xhq 
4XHQ 4xi5 
4XI5 4xmk 
4XMK 4xml 
4XML 4xmm 
4XMM 4xmn 
4XMN 4xmp 
4XMP 4xnm 
4XNM 4xnq 
4XNQ 4xnu 
4XNU 4xnx 
4XNX 4xny 
4XNY 4xnz 
4XNZ 4xp1 
4XP1 4xp4 
4XP4 4xp5 
4XP5 4xp6 
4XP6 4xp9 
4XP9 4xpa 
4XPA 4xpb 
4XPB 4xpf 
4XPF 4xpg 
4XPG 4xph 
4XPH 4xpt 
4XPT 4xrc 
4XRC 4xtr 
4XTR 4xvs 
4XVS 4xvt 
4XVT 4xvu 
4XVU 4xwg 
4XWG 4xwo 
4XWO 4xx1 
4XX1 4xxd 
4XXD 4xzu 
4XZU 4y16 
4Y16 4y19 
4Y19 4y1a 
4Y1A 4y2d 
4Y2D 4y4f 
4Y4F 4y4h 
4Y4H 4y4k 
4Y4K 4yaq 
4YAQ 4ybh 
4YBH 4ybl 
4YBL 4yc2 
4YC2 4ydi 
4YDI 4ydj 
4YDJ 4ydk 
4YDK 4ydl 
4YDL 4ydv 
4YDV 4ye4 
4YE4 4ye6 
4YE6 4ye8 
4YE8 4ye9 
4YE9 4yfc 
4YFC 4yfl 
4YFL 4ygv 
4YGV 4yh6 
4YH6 4yh7 
4YH7 4yhi 
4YHI 4yhk 
4YHK 4yhl 
4YHL 4yhm 
4YHM 4yhn 
4YHN 4yho 
4YHO 4yhp 
4YHP 4yhy 
4YHY 4yhz 
4YHZ 4yk4 
4YK4 4yny 
4YNY 4yo0 
4YO0 4ypg 
4YPG 4yr6 
4YR6 4yue 
4YUE 4ywg 
4YWG 4yx2 
4YX2 4yxh 
4YXH 4yxk 
4YXK 4yxl 
4YXL 4yzf 
4YZF 4z0b 
4Z0B 4z18 
4Z18 4z5r 
4Z5R 4z7n 
4Z7N 4z7o 
4Z7O 4z7q 
4Z7Q 4z7u 
4Z7U 4z7v 
4Z7V 4z7w 
4Z7W 4z8f 
4Z8F 4z95 
4Z95 4zak 
4ZAK 4zd3 
4ZD3 4zdh 
4ZDH 4zez 
4ZEZ 4zff 
4ZFF 4zfg 
4ZFG 4zfo 
4ZFO 4znc 
4ZNC 4zne 
4ZNE 4zpt 
4ZPT 4zpv 
4ZPV 4zqk 
4ZQK 4zs6 
4ZS6 4zs7 
4ZS7 4zso 
4ZSO 4zto 
4ZTO 4ztp 
4ZTP 4zxb 
4ZXB 4zyk 
4ZYK 4zyp 
4ZYP 5a16 
5A16 5a2f 
5A2F 5a3i 
5A3I 5a7x 
5A7X 5a8h 
5A8H 5aco 
5ACO 5ado 
5ADO 5adp 
5ADP 5alb 
5ALB 5alc 
5ALC 5anm 
5ANM 5any 
5ANY 5aum 
5AUM 5awn 
5AWN 5az2 
5AZ2 5aze 
5AZE 5b3j 
5B3J 5bmf 
5BMF 5bo1 
5BO1 5bq7 
5BQ7 5brz 
5BRZ 5bs0 
5BS0 5bv7 
5BV7 5bvj 
5BVJ 5bvp 
5BVP 5bw7 
5BW7 5bzd 
5BZD 5bzw 
5BZW 5c07 
5C07 5c08 
5C08 5c09 
5C09 5c0a 
5C0A 5c0b 
5C0B 5c0c 
5C0C 5c0n 
5C0N 5c0r 
5C0R 5c0s 
5C0S 5c3t 
5C3T 5c6t 
5C6T 5c7k 
5C7K 5c7x 
5C7X 5c8j 
5C8J 5cay 
5CAY 5cck 
5CCK 5cd3 
5CD3 5cd5 
5CD5 5cex 
5CEX 5cey 
5CEY 5cez 
5CEZ 5cgy 
5CGY 5chn 
5CHN 5cil 
5CIL 5cin 
5CIN 5cip 
5CIP 5cj8 
5CJ8 5cjb 
5CJB 5cjo 
5CJO 5cjq 
5CJQ 5cjs 
5CJS 5cjx 
5CJX 5cma 
5CMA 5cp3 
5CP3 5cp7 
5CP7 5csz 
5CSZ 5cus 
5CUS 5cws 
5CWS 5czv 
5CZV 5czx 
5CZX 5d1q 
5D1Q 5d1x 
5D1X 5d1z 
5D1Z 5d2l 
5D2L 5d2n 
5D2N 5d4q 
5D4Q 5d5m 
5D5M 5d6c 
5D6C 5d6d 
5D6D 5d70 
5D70 5d71 
5D71 5d72 
5D72 5d7i 
5D7I 5d7j 
5D7J 5d7k 
5D7K 5d7l 
5D7L 5d7s 
5D7S 5d8j 
5D8J 5d93 
5D93 5d96 
5D96 5d9q 
5D9Q 5dd0 
5DD0 5dd1 
5DD1 5dd3 
5DD3 5dd5 
5DD5 5dd6 
5DD6 5dfv 
5DFV 5di8 
5DI8 5dj0 
5DJ0 5dj2 
5DJ2 5dj6 
5DJ6 5dj8 
5DJ8 5dja 
5DJA 5djc 
5DJC 5djd 
5DJD 5djx 
5DJX 5djy 
5DJY 5djz 
5DJZ 5dk0 
5DK0 5dk2 
5DK2 5dk3 
5DK3 5dlm 
5DLM 5dmg 
5DMG 5dmi 
5DMI 5do2 
5DO2 5dq9 
5DQ9 5dqd 
5DQD 5dqj 
5DQJ 5dr5 
5DR5 5drn 
5DRN 5drw 
5DRW 5drx 
5DRX 5drz 
5DRZ 5ds8 
5DS8 5dsc 
5DSC 5dt1 
5DT1 5dtf 
5DTF 5dub 
5DUB 5dum 
5DUM 5dup 
5DUP 5dur 
5DUR 5dvk 
5DVK 5dvl 
5DVL 5dvm 
5DVM 5dvn 
5DVN 5dvo 
5DVO 5dwu 
5DWU 5dyo 
5DYO 5e08 
5E08 5e1a 
5E1A 5e2t 
5E2T 5e2u 
5E2U 5e2v 
5E2V 5e2w 
5E2W 5e4i 
5E4I 5e5r 
5E5R 5e5u 
5E5U 5e6i 
5E6I 5e8d 
5E8D 5e8e 
5E8E 5e94 
5E94 5e99 
5E99 5ea0 
5EA0 5ebl 
5EBL 5ebm 
5EBM 5ebw 
5EBW 5ec1 
5EC1 5ec2 
5EC2 5eii 
5EII 5eiq 
5EIQ 5eiv 
5EIV 5en2 
5EN2 5eo9 
5EO9 5eoc 
5EOC 5eoq 
5EOQ 5eor 
5EOR 5epm 
5EPM 5erw 
5ERW 5esa 
5ESA 5esq 
5ESQ 5esv 
5ESV 5esz 
5ESZ 5etu 
5ETU 5eu6 
5EU6 5eu7 
5EU7 5euk 
5EUK 5euo 
5EUO 5ewi 
5EWI 5f1d 
5F1D 5f3b 
5F3B 5f3h 
5F3H 5f6h 
5F6H 5f6i 
5F6I 5f6j 
5F6J 5f7e 
5F7E 5f88 
5F88 5f96 
5F96 5f9o 
5F9O 5f9w 
5F9W 5fb8 
5FB8 5fcu 
5FCU 5fec 
5FEC 5ff6 
5FF6 5fgb 
5FGB 5fgc 
5FGC 5fha 
5FHA 5fhb 
5FHB 5fhc 
5FHC 5fhx 
5FHX 5fk9 
5FK9 5fka 
5FKA 5fuo 
5FUO 5fuu 
5FUU 5fuz 
5FUZ 5fyj 
5FYJ 5fyk 
5FYK 5fyl 
5FYL 5ggq 
5GGQ 5ggr 
5GGR 5ggs 
5GGS 5ggt 
5GGT 5ggu 
5GGU 5ggv 
5GGV 5ghw 
5GHW 5gir 
5GIR 5gis 
5GIS 5grj 
5GRJ 5gzr 
5GZR 5hcg 
5HCG 5hdb 
5HDB 5hdq 
5HDQ 5hhm 
5HHM 5hho 
5HHO 5hhv 
5HHV 5hhx 
5HHX 5hi3 
5HI3 5hi4 
5HI4 5hi5 
5HI5 5hj3 
5HJ3 5hpm 
5HPM 5hvw 
5HVW 5hyj 
5HYJ 5hyq 
5HYQ 5hys 
5HYS 5i15 
5I15 5i16 
5I16 5i17 
5I17 5i18 
5I18 5i19 
5I19 5i1a 
5I1A 5i1c 
5I1C 5i1d 
5I1D 5i1e 
5I1E 5i1g 
5I1G 5i1h 
5I1H 5i1i 
5I1I 5i1j 
5I1J 5i1k 
5I1K 5i1l 
5I1L 5i2i 
5I2I 5i30 
5I30 5i5k 
5I5K 5i66 
5I66 5i6x 
5I6X 5i6z 
5I6Z 5i71 
5I71 5i73 
5I73 5i74 
5I74 5i75 
5I75 5i8c 
5I8C 5i8e 
5I8E 5i8h 
5I8H 5i8k 
5I8K 5i8o 
5I8O 5i99 
5I99 5i9q 
5I9Q 5ibl 
5IBL 5ibt 
5IBT 5ibu 
5IBU 5icx 
5ICX 5icy 
5ICY 5icz 
5ICZ 5id0 
5ID0 5id1 
5ID1 5ies 
5IES 5if0 
5IF0 5ifa 
5IFA 5igx 
5IGX 5ihu 
5IHU 5ijv 
5IJV 5ikc 
5IKC 5ilt 
5ILT 5iop 
5IOP 5iq7 
5IQ7 5iq9 
5IQ9 5ir1 
5IR1 5itb 
5ITB 5itf 
5ITF 5ius 
5IUS 5iv2 
5IV2 5ivz 
5IVZ 5iwl 
5IWL 5j3d 
5J3D 5j89 
5J89 5j8o 
5J8O 5j9p 
5J9P 5jhl 
5JHL 5jny 
5JNY 5jo5 
5JO5 5jof 
5JOF 5jr1 
5JR1 5js9 
5JS9 5jsa 
5JSA 5jue 
5JUE 5jw3 
5JW3 5jw4 
5JW4 5jw5 
5JW5 5jxa 
5JXA 5jxe 
5JXE 5k59 
5K59 5k6u 
5K6U 5k6v 
5K6V 5k6w 
5K6W 5k6x 
5K6X 5k6y 
5K6Y 5k6z 
5K6Z 5k70 
5K70 5k9j 
5K9J 5k9q 
5K9Q 5kan 
5KAN 5kjr 
5KJR 5kn5 
5KN5 5ks9 
5KS9 5ksa 
5KSA 5ksb 
5KSB 5kte 
5KTE 5kvd 
5KVD 5kvf 
5KVF 5kvg 
5KVG 5kw9 
5KW9 5kzc 
5KZC 5l0q 
5L0Q 5l2k 
5L2K 5l7x 
5L7X 5l88 
5L88 5l9d 
5L9D 5lcv 
5LCV 5lgh 
5LGH 5sws 
5SWS 5swz 
5SWZ 5sx4 
5SX4 5sx5 
5SX5 5t1k 
5T1K 5t1l 
5T1L 5t1m 
5T1M 5t33 
5T33 5t3s 
5T3S 5t3x 
5T3X 5t3z 
5T3Z 5t5f 
5T5F 5t5n 
5T5N 5te4 
5TE4 5te6 
5TE6 5te7 
5TE7 5tf1 
5TF1 5tfs 
5TFS 5tgb 
5TGB 5th2 
5TH2 5thr 
5THR 6fab 
THREE-DIMENSIONAL STRUCTURE OF MURINE ANTI-P-AZOPHENYLARSONATE FAB 36-71. 1. X-RAY CRYSTALLOGRAPHY, SITE-DIRECTED MUTAGENESIS, AND MODELING OF THE COMPLEX WITH HAPTEN 7fab 
CRYSTAL STRUCTURE OF HUMAN IMMUNOGLOBULIN FRAGMENT FAB NEW REFINED AT 2.0 ANGSTROMS RESOLUTION 8fab 
CRYSTAL STRUCTURE OF THE FAB FRAGMENT FROM THE HUMAN MYELOMA IMMUNOGLOBULIN IGG HIL AT 1.8 ANGSTROMS RESOLUTION

