The domain within your query sequence starts at position 286 and ends at position 356; the E-value for the KH domain shown below is 1.22e-2.
IDEQLRQIGSRSYSGRGRGRRGPNYTSGYGTNSELSNPSETESERKDELSDWSLAGEDDR ETRHQRDSRRR
KHK homology RNA-binding domain |
|---|
| SMART accession number: | SM00322 |
|---|---|
| Description: | - |
| Interpro abstract (IPR004087): | The K homology (KH) domain was first identified in the human heterogeneous nuclear ribonucleoprotein (hnRNP) K. An evolutionarily conserved sequence of around 70 amino acids, the KH domain is present in a wide variety of nucleic acid-binding proteins. The KH domain binds RNA, and can function in RNA recognition [ (PUBMED:17437720) ]. It is found in multiple copies in several proteins, where they can function cooperatively or independently. For example, in the AU-rich element RNA-binding protein KSRP, which has 4 KH domains, KH domains 3 and 4 behave as independent binding modules to interact with different regions of the AU-rich RNA targets [ (PUBMED:17437720) ]. The solution structure of the first KH domain of FMR1 [ (PUBMED:9302998) ] and of the C-terminal KH domain of hnRNP K [ (PUBMED:10369774) ] determined by nuclear magnetic resonance (NMR) revealed a beta-alpha-alpha-beta-beta-alpha structure. Proteins containing KH domains include:
According to structural analyses [ (PUBMED:9302998) (PUBMED:10369774) (PUBMED:11160884) ], the KH domain can be separated in two groups - type 1 and type 2. |
| GO function: | nucleic acid binding (GO:0003676) |
| Family alignment: |
There are 181505 KH domains in 115166 proteins in SMART's nrdb database.
Click on the following links for more information.
- Evolution (species in which this domain is found)
-
Taxonomic distribution of proteins containing KH domain.
This tree includes only several representative species. The complete taxonomic breakdown of all proteins with KH domain is also avaliable.
Click on the protein counts, or double click on taxonomic names to display all proteins containing KH domain in the selected taxonomic class.
- Cellular role (predicted cellular role)
-
Binding / catalysis: RNA-binding
- Literature (relevant references for this domain)
-
Primary literature is listed below; Automatically-derived, secondary literature is also avaliable.
- Lewis HA et al.
- Sequence-specific RNA binding by a Nova KH domain: implications for paraneoplastic disease and the fragile X syndrome.
- Cell. 2000; 100: 323-32
- Display abstract
The structure of a Nova protein K homology (KH) domain recognizing single-stranded RNA has been determined at 2.4 A resolution. Mammalian Nova antigens (1 and 2) constitute an important family of regulators of RNA metabolism in neurons, first identified using sera from cancer patients with the autoimmune disorder paraneoplastic opsoclonus-myoclonus ataxia (POMA). The structure of the third KH domain (KH3) of Nova-2 bound to a stem loop RNA resembles a molecular vise, with 5'-Ura-Cyt-Ade-Cyt-3' pinioned between an invariant Gly-X-X-Gly motif and the variable loop. Tetranucleotide recognition is supported by an aliphatic alpha helix/beta sheet RNA-binding platform, which mimics 5'-Ura-Gua-3' by making Watson-Crick-like hydrogen bonds with 5'-Cyt-Ade-3'. Sequence conservation suggests that fragile X mental retardation results from perturbation of RNA binding by the FMR1 protein.
- Adinolfi S, Bagni C, Musco G, Gibson T, Mazzarella L, Pastore A
- Dissecting FMR1, the protein responsible for fragile X syndrome, in its structural and functional domains.
- RNA. 1999; 5: 1248-58
- Display abstract
FMR1 is an RNA-binding protein that is either absent or mutated in patients affected by the fragile X syndrome, the most common inherited cause of mental retardation in humans. Sequence analysis of the FMR1 protein has suggested that RNA binding is related to the presence of two K-homologous (KH) modules and an RGG box. However, no attempt has been so far made to map the RNA-binding sites along the protein sequence and to identify possible differential RNA-sequence specificity. In the present article, we describe work done to dissect FMR1 into regions with structurally and functionally distinct properties. A semirational approach was followed to identify four regions: an N-terminal stretch of 200 amino acids, the two KH regions, and a C-terminal stretch. Each region was produced as a recombinant protein, purified, and probed for its state of folding by spectroscopical techniques. Circular dichroism and NMR spectra of the N-terminus show formation of secondary structure with a strong tendency to aggregate. Of the two homologous KH motifs, only the first one is folded whereas the second remains unfolded even when it is extended both N- and C-terminally. The C-terminus is, as expected from its amino acid composition, nonglobular. Binding assays were then performed using the 4-nt homopolymers. Our results show that only the first KH domain but not the second binds to RNA, and provide the first direct evidence for RNA binding of both the N-terminal and the C-terminal regions. RNA binding for the N-terminus could not be predicted from sequence analysis because no known RNA-binding motif is identifiable in this region. Different sequence specificity was observed for the fragments: both the N-terminus of the protein and KH1 bind preferentially to poly-(rG). The C-terminal region, which contains the RGG box, is nonspecific, as it recognizes the bases with comparable affinity. We therefore conclude that FMR1 is a protein with multiple sites of interaction with RNA: sequence specificity is most likely achieved by the whole block that comprises the first approximately 400 residues, whereas the C-terminus provides a nonspecific binding surface.
- Ostareck-Lederer A, Ostareck DH, Hentze MW
- Cytoplasmic regulatory functions of the KH-domain proteins hnRNPs K and E1/E2.
- Trends Biochem Sci. 1998; 23: 409-11
- Musco G et al.
- The solution structure of the first KH domain of FMR1, the protein responsible for the fragile X syndrome.
- Nat Struct Biol. 1997; 4: 712-6
- Musco G et al.
- Three-dimensional structure and stability of the KH domain: molecular insights into the fragile X syndrome.
- Cell. 1996; 85: 237-45
- Display abstract
The KH module is a sequence motif found in a number of proteins that are known to be in close association with RNA. Experimental evidence suggests a direct involvement of KH in RNA binding. The human FMR1 protein, which has two KH domains, is associated with fragile X syndrome, the most common inherited cause of mental retardation. Here we present the three-dimensional solution structure of the KH module. The domain consists of a stable beta alpha alpha beta beta alpha fold. On the basis of our results, we suggest a potential surface for RNA binding centered on the loop between the first two helices. Substitution of a well-conserved hydrophobic residue located on the second helix destroys the KH fold; a mutation of this position in FMR1 leads to an aggravated fragile X phenotype.
- Siomi H, Choi M, Siomi MC, Nussbaum RL, Dreyfuss G
- Essential role for KH domains in RNA binding: impaired RNA binding by a mutation in the KH domain of FMR1 that causes fragile X syndrome.
- Cell. 1994; 77: 33-9
- Display abstract
The KH domain is an evolutionarily conserved sequence motif present in many RNA-binding proteins, including the pre-mRNA-binding (hnRNP) K protein and the fragile X mental retardation gene product (FMR1). We assessed the role of KH domains in RNA binding by mutagenesis of KH domains in hnRNP K and FMR1. Conserved residues of all three hnRNP K KH domains are required for its wild-type RNA binding. Interestingly, while fragile X syndrome is usually caused by lack of FMR1 expression, a previously reported mutation in a highly conserved residue of one of its two KH domains (Ile-304-->Asn) also results in mental retardation. We found that the binding of this mutant protein to RNA is severely impaired. These results demonstrate an essential role for KH domains in RNA binding. Furthermore, they strengthen the connection between fragile X syndrome and loss of the RNA binding activity of FMR1.
- Disease (disease genes where sequence variants are found in this domain)
-
SwissProt sequences and OMIM curated human diseases associated with missense mutations within the KH domain.
Protein Disease Synaptic functional regulator FMR1 (Q06787) (SMART) OMIM:309550: Fragile X syndrome - Metabolism (metabolic pathways involving proteins which contain this domain)
-
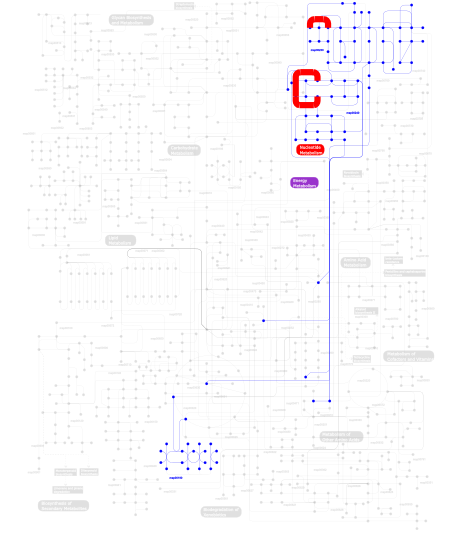
Click the image to view the interactive version of the map in iPath% proteins involved KEGG pathway ID Description 34.62  map00240
map00240Pyrimidine metabolism 34.62  map00230
map00230Purine metabolism 29.68 map03010 Ribosome 1.00 map02010 ABC transporters - General 0.08  map00190
map00190Oxidative phosphorylation This information is based on mapping of SMART genomic protein database to KEGG orthologous groups. Percentage points are related to the number of proteins with KH domain which could be assigned to a KEGG orthologous group, and not all proteins containing KH domain. Please note that proteins can be included in multiple pathways, ie. the numbers above will not always add up to 100%.
- Structure (3D structures containing this domain)
3D Structures of KH domains in PDB
PDB code Main view Title 1dt4 
CRYSTAL STRUCTURE OF NOVA-1 KH3 K-HOMOLOGY RNA-BINDING DOMAIN 1dtj 
CRYSTAL STRUCTURE OF NOVA-2 KH3 K-HOMOLOGY RNA-BINDING DOMAIN 1e3h 
SeMet derivative of Streptomyces antibioticus PNPase/GPSI enzyme 1e3p 
tungstate derivative of Streptomyces antibioticus PNPase/GPSI enzyme 1ec6 
CRYSTAL STRUCTURE OF NOVA-2 KH3 K-HOMOLOGY RNA-BINDING DOMAIN BOUND TO 20-MER RNA HAIRPIN 1fjg 
STRUCTURE OF THE THERMUS THERMOPHILUS 30S RIBOSOMAL SUBUNIT IN COMPLEX WITH THE ANTIBIOTICS STREPTOMYCIN, SPECTINOMYCIN, AND PAROMOMYCIN 1hh2 
Crystal structure of NusA from Thermotoga maritima 1hnw 
STRUCTURE OF THE THERMUS THERMOPHILUS 30S RIBOSOMAL SUBUNIT IN COMPLEX WITH TETRACYCLINE 1hnx 
STRUCTURE OF THE THERMUS THERMOPHILUS 30S RIBOSOMAL SUBUNIT IN COMPLEX WITH PACTAMYCIN 1hnz 
STRUCTURE OF THE THERMUS THERMOPHILUS 30S RIBOSOMAL SUBUNIT IN COMPLEX WITH HYGROMYCIN B 1hr0 
CRYSTAL STRUCTURE OF INITIATION FACTOR IF1 BOUND TO THE 30S RIBOSOMAL SUBUNIT 1i94 
CRYSTAL STRUCTURES OF THE SMALL RIBOSOMAL SUBUNIT WITH TETRACYCLINE, EDEINE AND IF3 1i95 
CRYSTAL STRUCTURE OF THE 30S RIBOSOMAL SUBUNIT FROM THERMUS THERMOPHILUS IN COMPLEX WITH EDEINE 1i96 
CRYSTAL STRUCTURE OF THE 30S RIBOSOMAL SUBUNIT FROM THERMUS THERMOPHILUS IN COMPLEX WITH THE TRANSLATION INITIATION FACTOR IF3 (C-TERMINAL DOMAIN) 1i97 
CRYSTAL STRUCTURE OF THE 30S RIBOSOMAL SUBUNIT FROM THERMUS THERMOPHILUS IN COMPLEX WITH TETRACYCLINE 1ibk 
STRUCTURE OF THE THERMUS THERMOPHILUS 30S RIBOSOMAL SUBUNIT IN COMPLEX WITH THE ANTIBIOTIC PAROMOMYCIN 1ibl 
STRUCTURE OF THE THERMUS THERMOPHILUS 30S RIBOSOMAL SUBUNIT IN COMPLEX WITH A MESSENGER RNA FRAGMENT AND COGNATE TRANSFER RNA ANTICODON STEM-LOOP BOUND AT THE A SITE AND WITH THE ANTIBIOTIC PAROMOMYCIN 1ibm 
STRUCTURE OF THE THERMUS THERMOPHILUS 30S RIBOSOMAL SUBUNIT IN COMPLEX WITH A MESSENGER RNA FRAGMENT AND COGNATE TRANSFER RNA ANTICODON STEM-LOOP BOUND AT THE A SITE 1j4w 
COMPLEX OF THE KH3 and KH4 DOMAINS OF FBP WITH A SINGLE_STRANDED 29mer DNA OLIGONUCLEOTIDE FROM THE FUSE ELEMENT OF THE C-MYC ONCOGENE 1j5e 
Structure of the Thermus thermophilus 30S Ribosomal Subunit 1j5k 
COMPLEX OF THE KH3 DOMAIN OF HNRNP K WITH A SINGLE_STRANDED 10MER DNA OLIGONUCLEOTIDE 1jgo 
The Path of Messenger RNA Through the Ribosome. THIS FILE, 1JGO, CONTAINS THE 30S RIBOSOME SUBUNIT, THREE TRNA, AND MRNA MOLECULES. 50S RIBOSOME SUBUNIT IS IN THE FILE 1GIY 1jgp 
The Path of Messenger RNA Through the Ribosome. THIS FILE, 1JGP, CONTAINS THE 30S RIBOSOME SUBUNIT, THREE TRNA, AND MRNA MOLECULES. 50S RIBOSOME SUBUNIT IS IN THE FILE 1GIY 1jgq 
The Path of Messenger RNA Through the Ribosome. THIS FILE, 1JGQ, CONTAINS THE 30S RIBOSOME SUBUNIT, THREE TRNA, AND MRNA MOLECULES. 50S RIBOSOME SUBUNIT IS IN THE FILE 1GIY 1k0r 
Crystal Structure of Mycobacterium tuberculosis NusA 1k1g 
STRUCTURAL BASIS FOR RECOGNITION OF THE INTRON BRANCH SITE RNA BY SPLICING FACTOR 1 1khm 
C-TERMINAL KH DOMAIN OF HNRNP K (KH3) 1l2f 
Crystal structure of NusA from Thermotoga maritima: a structure-based role of the N-terminal domain 1ml5 
Structure of the E. coli ribosomal termination complex with release factor 2 1n32 
Structure of the Thermus thermophilus 30S ribosomal subunit bound to codon and near-cognate transfer RNA anticodon stem-loop mismatched at the first codon position at the a site with paromomycin 1n33 
Structure of the Thermus thermophilus 30S ribosomal subunit bound to codon and near-cognate transfer rna anticodon stem-loop mismatched at the second codon position at the a site with paromomycin 1n34 
Structure of the Thermus thermophilus 30S ribosomal subunit in the presence of codon and crystallographically disordered near-cognate transfer rna anticodon stem-loop mismatched at the first codon position 1n36 
Structure of the Thermus thermophilus 30S ribosomal subunit in the presence of crystallographically disordered codon and near-cognate transfer RNA anticodon stem-loop mismatched at the second codon position 1tua 
1.5 A Crystal Structure of a Protein of Unknown Function APE0754 from Aeropyrum pernix 1vig 
NMR STUDY OF VIGILIN, REPEAT 6, 40 STRUCTURES 1vih 
NMR STUDY OF VIGILIN, REPEAT 6, MINIMIZED AVERAGE STRUCTURE 1vvj 
1VVJ 1vy4 
1VY4 1vy5 
1VY5 1vy6 
1VY6 1vy7 
1VY7 1we8 
Solution structure of KH domain in protein BAB28342 1wvn 
Crsytal Structure of domain 3 of human alpha polyC binding protein 1x4m 
Solution structure of KH domain in Far upstream element binding protein 1 1x4n 
Solution structure of KH domain in FUSE binding protein 1 1xmo 
Crystal Structure of mnm5U34t6A37-tRNALysUUU Complexed with AAG-mRNA in the Decoding Center 1xmq 
Crystal Structure of t6A37-ASLLysUUU AAA-mRNA Bound to the Decoding Center 1xnq 
Structure of an Inosine-Adenine Wobble Base Pair Complex in the Context of the Decoding Center 1xnr 
Crystal Structure of an Inosine-Cytosine Wobble Base Pair in the Context of the Decoding Center 1ztg 
human alpha polyC binding protein KH1 1zzi 
Crystal Structure Analysis of the third KH domain of hnRNP K in complex with ssDNA 1zzj 
Structure of the third KH domain of hnRNP K in complex with 15-mer ssDNA 1zzk 
Crystal Structure of the third KH domain of hnRNP K at 0.95A resolution 2ann 
Crystal structure (I) of Nova-1 KH1/KH2 domain tandem with 25 nt RNA hairpin 2anr 
Crystal structure (II) of Nova-1 KH1/KH2 domain tandem with 25nt RNA hairpin 2asb 
Structure of a Mycobacterium tuberculosis NusA-RNA complex 2atw 
Structure of a Mycobacterium tuberculosis NusA-RNA complex 2axy 
Crystal Structure of KH1 domain of human Poly(C)-binding protein-2 with C-rich strand of human telomeric DNA 2ba0 
Archaeal exosome core 2bl5 
Solution structure of the KH-QUA2 region of the Xenopus STAR-GSG Quaking protein. 2cpq 
Solution structure of the N-terminal KH domain of human FXR1 2cte 
Solution structure of the 1st KH type I domain from human Vigilin 2ctf 
Solution structure of the 4th KH type I domain from human Vigilin 2ctj 
Solution structure of the 8th KH type I domain from human Vigilin 2ctk 
Solution structure of the 12th KH type I domain from human Vigilin 2ctl 
Solution structure of the 13th KH type I domain from human Vigilin 2ctm 
Solution structure of the 14th KH type I domain from human Vigilin 2cxc 
Crystal structure of archaeal transcription termination factor NusA 2cy1 
Crystal structure of APE1850 2dgr 
Solution structure of the second KH domain in ring finger and KH domain containing protein 1 2e3u 
Crystal structure analysis of Dim2p from Pyrococcus horikoshii OT3 2e5l 
A snapshot of the 30S ribosomal subunit capturing mRNA via the Shine- Dalgarno interaction 2f4v 
30S ribosome + designer antibiotic 2fmr 
KH1 FROM THE FRAGILE X PROTEIN FMR1, NMR, 18 STRUCTURES 2hh2 
Solution structure of the fourth KH domain of KSRP 2hh3 
Solution structure of the third KH domain of KSRP 2hhh 
Crystal structure of kasugamycin bound to the 30S ribosomal subunit 2je6 
Structure of a 9-subunit archaeal exosome 2jea 
Structure of a 9-subunit archaeal exosome bound to RNA 2jeb 
Structure of a 9-subunit archaeal exosome bound to Mn ions 2jvz 
Solution NMR Structure of the Second and Third KH Domains of KSRP 2jzx 
PCBP2 KH1-KH2 domains 2mjh 
2MJH 2opu 
Solution NMR Structure of the First Domain of KSRP 2opv 
Solution NMR Structure of the Second Domain of KSRP 2p2r 
Crystal structure of the third KH domain of human Poly(C)-Binding Protein-2 in complex with C-rich strand of human telomeric DNA 2pqu 
Crystal structure of KH1 domain of human PCBP2 complexed to single-stranded 12-mer telomeric dna 2py9 
Protein-RNA Interaction involving KH1 domain from Human Poly(C)-Binding Protein-2 2qnd 
Crystal Structure of the KH1-KH2 domains from human Fragile X Mental Retardation Protein 2uu9 
Structure of the Thermus thermophilus 30S ribosomal subunit complexed with a Valine-ASL with cmo5U in position 34 bound to an mRNA with a GUG-codon in the A-site and paromomycin. 2uua 
Structure of the Thermus thermophilus 30S ribosomal subunit complexed with a Valine-ASL with cmo5U in position 34 bound to an mRNA with a GUC-codon in the A-site and paromomycin. 2uub 
Structure of the Thermus thermophilus 30S ribosomal subunit complexed with a Valine-ASL with cmo5U in position 34 bound to an mRNA with a GUU-codon in the A-site and paromomycin. 2uuc 
Structure of the Thermus thermophilus 30S ribosomal subunit complexed with a Valine-ASL with cmo5U in position 34 bound to an mRNA with a GUA-codon in the A-site and paromomycin. 2uxb 
Crystal structure of an extended tRNA anticodon stem loop in complex with its cognate mRNA GGGU in the context of the Thermus thermophilus 30S subunit. 2uxc 
Crystal structure of an extended tRNA anticodon stem loop in complex with its cognate mRNA UCGU in the context of the Thermus thermophilus 30S subunit. 2uxd 
Crystal structure of an extended tRNA anticodon stem loop in complex with its cognate mRNA CGGG in the context of the Thermus thermophilus 30S subunit. 2vqe 
Modified uridines with C5-methylene substituents at the first position of the tRNA anticodon stabilize U-G wobble pairing during decoding 2vqf 
Modified uridines with C5-methylene substituents at the first position of the tRNA anticodon stabilize U-G wobble pairing during decoding 2xr1 
DIMERIC ARCHAEAL CLEAVAGE AND POLYADENYLATION SPECIFICITY FACTOR WITH N-TERMINAL KH DOMAINS (KH-CPSF) FROM METHANOSARCINA MAZEI 2ycb 
Structure of the archaeal beta-CASP protein with N-terminal KH domains from Methanothermobacter thermautotrophicus 2ykr 
30S ribosomal subunit with RsgA bound in the presence of GMPPNP 2z0s 
Crystal structure of putative exosome complex RNA-binding protein 2zm6 
Crystal structure of the Thermus thermophilus 30S ribosomal subunit 3aev 
Crystal structure of a/eIF2alpha-aDim2p-rRNA complex from Pyrococcus horikoshii OT3 3af5 
The crystal structure of an archaeal CPSF subunit, PH1404 from Pyrococcus horikoshii 3af6 
The crystal structure of an archaeal CPSF subunit, PH1404 from Pyrococcus horikoshii complexed with RNA-analog 3cdi 
Crystal structure of E. coli PNPase 3gku 
Crystal structure of a probable RNA-binding protein from Clostridium symbiosum ATCC 14940 3j6x 
3J6X 3j6y 
3J6Y 3j77 
3J77 3j78 
3J78 3j7a 
3J7A 3j7p 
3J7P 3j7r 
3J7R 3j80 
3J80 3j81 
3J81 3j9w 
3J9W 3j9y 
3J9Y 3j9z 
3J9Z 3ja1 
3JA1 3jag 
3JAG 3jah 
3JAH 3jai 
3JAI 3jaj 
3JAJ 3jam 
3JAM 3jan 
3JAN 3jap 
3JAP 3jaq 
3JAQ 3jbn 
3JBN 3jbo 
3JBO 3jbp 
3JBP 3jbu 
3JBU 3jbv 
3JBV 3jcd 
3JCD 3jce 
3JCE 3jcj 
3JCJ 3jcn 
3JCN 3krm 
Imp1 kh34 3l7z 
Crystal structure of the S. solfataricus archaeal exosome 3oto 
Crystal Structure of the 30S ribosomal subunit from a KsgA mutant of Thermus thermophilus (HB8) 3t1h 
Structure of the Thermus thermophilus 30S ribosomal subunit complexed with a human anti-codon stem loop (HASL) of transfer RNA lysine 3 (tRNALys3) bound to an mRNA with an AAA-codon in the A-site and Paromomycin 3t1y 
Structure of the Thermus thermophilus 30S ribosomal subunit complexed with a human anti-codon stem loop (HASL) of transfer RNA Lysine 3 (TRNALYS3) bound to an mRNA with an AAG-codon in the A-site and paromomycin 3u1k 
Crystal structure of human PNPase 3vke 
Contribution of the first K-homology domain of poly(C)-binding protein 1 to its affinity and specificity for C-rich oligonucleotides 4a2i 
Cryo-electron Microscopy Structure of the 30S Subunit in Complex with the YjeQ Biogenesis Factor 4adv 
Structure of the E. coli methyltransferase KsgA bound to the E. coli 30S ribosomal subunit 4aid 
Crystal structure of C. crescentus PNPase bound to RNase E recognition peptide 4aim 
Crystal structure of C. crescentus PNPase bound to RNase E recognition peptide 4am3 
Crystal structure of C. crescentus PNPase bound to RNA 4aqy 
Structure of ribsome-apramycin complexes 4b3m 
Crystal structure of the 30S ribosome in complex with compound 1 4b3r 
Crystal structure of the 30S ribosome in complex with compound 30 4b3s 
Crystal structure of the 30S ribosome in complex with compound 37 4b3t 
Crystal structure of the 30S ribosome in complex with compound 39 4b8t 
RNA BINDING PROTEIN Solution structure of the third KH domain of KSRP in complex with the G-rich target sequence. 4ba1 
Archaeal exosome (Rrp4-Rrp41(D182A)-Rrp42) bound to inorganic phosphate 4ba2 
Archaeal exosome (Rrp4-Rrp41(D182A)-Rrp42) bound to inorganic phosphate 4bsz 
Crystal Structure of the Yeast Ribosomal Protein Rps3 in Complex with its Chaperone Yar1 4d5l 
4D5L 4d61 
4D61 4dr1 
Crystal structure of the apo 30S ribosomal subunit from Thermus thermophilus (HB8) 4dr2 
Crystal structure of the Thermus thermophilus (HB8) 30S ribosomal subunit with multiple copies of paromomycin molecules bound 4dr3 
Crystal structure of the Thermus thermophilus (HB8) 30S ribosomal subunit with streptomycin bound 4dr4 
Crystal structure of the Thermus thermophilus (HB8) 30S ribosomal subunit with codon, cognate transfer RNA anticodon stem-loop and multiple copies of paromomycin molecules bound 4dr5 
Crystal structure of the Thermus thermophilus (HB8) 30S ribosomal subunit with codon, crystallographically disordered cognate transfer RNA anticodon stem-loop and streptomycin bound 4dr6 
Crystal structure of the Thermus thermophilus (HB8) 30S ribosomal subunit with codon, near-cognate transfer RNA anticodon stem-loop mismatched at the first codon position and streptomycin bound 4dr7 
Crystal structure of the Thermus thermophilus (HB8) 30S ribosomal subunit with codon, crystallographically disordered near-cognate transfer RNA anticodon stem-loop mismatched at the second codon position, and streptomycin bound 4duy 
Crystal structure of the Thermus thermophilus 30S ribosomal subunit with a 16S rRNA mutation, U13C 4duz 
Crystal structure of the Thermus thermophilus 30S ribosomal subunit with a 16S rRNA mutation, U13C, bound with streptomycin 4dv0 
Crystal structure of the Thermus thermophilus 30S ribosomal subunit with a 16S rRNA mutation, U20G 4dv1 
Crystal structure of the Thermus thermophilus 30S ribosomal subunit with a 16S rRNA mutation, U20G, bound with streptomycin 4dv2 
Crystal structure of the Thermus thermophilus 30S ribosomal subunit with a 16S rRNA mutation, C912A 4dv3 
Crystal structure of the Thermus thermophilus 30S ribosomal subunit with a 16S rRNA mutation, C912A, bound with streptomycin 4dv4 
Crystal structure of the Thermus thermophilus 30S ribosomal subunit with a 16S rRNA mutation, A914G 4dv5 
Crystal structure of the Thermus thermophilus 30S ribosomal subunit with a 16S rRNA mutation, A914G, bound with streptomycin 4dv6 
Crystal structure of the Thermus thermophilus 30S ribosomal subunit with a 16S rRNA mutation, A915G 4dv7 
Crystal structure of the Thermus thermophilus 30S ribosomal subunit with a 16S rRNA mutation, A915G, bound with streptomycin 4gkj 
Structure of the Thermus thermophilus 30S ribosomal subunit complexed with a human mitochondrial anticodon stem loop (ASL) of transfer RNA Methionine (TRNAMET) bound to an mRNA with an AUG-codon in the A-site and paromomycin. 4gkk 
Structure of the Thermus thermophilus 30S ribosomal subunit complexed with a human mitochondrial anticodon stem loop (ASL) of transfer RNA Methionine (TRNAMET) bound to an mRNA with an AUA-codon in the A-site and paromomycin 4ji0 
Crystal Structure of 30S ribosomal subunit from Thermus thermophilus 4ji1 
Crystal Structure of 30S ribosomal subunit from Thermus thermophilus 4ji2 
Crystal Structure of 30S ribosomal subunit from Thermus thermophilus 4ji3 
Crystal Structure of 30S ribosomal subunit from Thermus thermophilus 4ji4 
Crystal Structure of 30S ribosomal subunit from Thermus thermophilus 4ji5 
Crystal Structure of 30S ribosomal subunit from Thermus thermophilus 4ji6 
Crystal Structure of 30S ribosomal subunit from Thermus thermophilus 4ji7 
Crystal Structure of 30S ribosomal subunit from Thermus thermophilus 4ji8 
Crystal Structure of 30S ribosomal subunit from Thermus thermophilus 4jv5 
Crystal structures of pseudouridinilated stop codons with ASLs 4jvh 
Structure of the star domain of quaking protein in complex with RNA 4jvy 
Structure of the STAR (signal transduction and activation of RNA) domain of GLD-1 bound to RNA 4jya 
Crystal structures of pseudouridinilated stop codons with ASLs 4k0k 
Crystal structure of the Thermus thermophilus 30S ribosomal subunit complexed with a serine-ASL and mRNA containing a stop codon 4khp 
Structure of the Thermus thermophilus 30S ribosomal subunit in complex with de-6-MSA-pactamycin 4kvb 
4KVB 4kzx 
Rabbit 40S ribosomal subunit in complex with eIF1. 4kzy 
Rabbit 40S ribosomal subunit in complex with eIF1 and eIF1A. 4kzz 
Rabbit 40S ribosomal subunit in complex with mRNA, initiator tRNA and eIF1A 4l47 
4L47 4l71 
4L71 4lel 
4LEL 4lf4 
4LF4 4lf5 
4LF5 4lf6 
4LF6 4lf7 
4LF7 4lf8 
4LF8 4lf9 
4LF9 4lfa 
4LFA 4lfb 
4LFB 4lfc 
4LFC 4lfz 
4LFZ 4lij 
Crystal structure of a far upstream element (FUSE) binding protein 1 (FUBP1) from Homo sapiens at 1.95 A resolution 4lnt 
4LNT 4lsk 
4LSK 4lt8 
4LT8 4mtn 
Crystal structure of transcription termination factor NusA from Planctomyces limnophilus DSM 3776 4nbq 
4NBQ 4nxm 
Crystal Structure of the 30S ribosomal subunit from a GidB (RsmG) mutant of Thermus thermophilus (HB8) 4nxn 
Crystal Structure of the 30S ribosomal subunit from a GidB (RsmG) mutant of Thermus thermophilus (HB8), bound with streptomycin 4ox9 
Crystal structure of the aminoglycoside resistance methyltransferase NpmA bound to the 30S ribosomal subunit 4p6f 
4P6F 4p70 
4P70 4qmf 
4QMF 4tua 
4TUA 4tub 
4TUB 4tuc 
4TUC 4tud 
4TUD 4tue 
4TUE 4u1u 
4U1U 4u1v 
4U1V 4u20 
4U20 4u24 
4U24 4u25 
4U25 4u26 
4U26 4u27 
4U27 4u3m 
4U3M 4u3n 
4U3N 4u3u 
4U3U 4u4n 
4U4N 4u4o 
4U4O 4u4q 
4U4Q 4u4r 
4U4R 4u4u 
4U4U 4u4y 
4U4Y 4u4z 
4U4Z 4u50 
4U50 4u51 
4U51 4u52 
4U52 4u53 
4U53 4u55 
4U55 4u56 
4U56 4u6f 
4U6F 4uer 
4UER 4ug0 
4UG0 4ujc 
4UJC 4ujd 
4UJD 4uje 
4UJE 4v3p 
4V3P 4v42 
4V42 4v47 
4V47 4v48 
4V48 4v49 
4V49 4v4a 
4V4A 4v4b 
4V4B 4v4g 
4V4G 4v4h 
4V4H 4v4i 
4V4I 4v4j 
4V4J 4v4n 
4V4N 4v4p 
4V4P 4v4q 
4V4Q 4v4r 
4V4R 4v4s 
4V4S 4v4t 
4V4T 4v4v 
4V4V 4v4w 
4V4W 4v4x 
4V4X 4v4y 
4V4Y 4v4z 
4V4Z 4v50 
4V50 4v51 
4V51 4v52 
4V52 4v53 
4V53 4v54 
4V54 4v55 
4V55 4v56 
4V56 4v57 
4V57 4v5a 
4V5A 4v5b 
4V5B 4v5c 
4V5C 4v5d 
4V5D 4v5e 
4V5E 4v5f 
4V5F 4v5g 
4V5G 4v5h 
4V5H 4v5j 
4V5J 4v5k 
4V5K 4v5l 
4V5L 4v5m 
4V5M 4v5n 
4V5N 4v5p 
4V5P 4v5q 
4V5Q 4v5r 
4V5R 4v5s 
4V5S 4v5y 
4V5Y 4v5z 
4V5Z 4v63 
4V63 4v64 
4V64 4v65 
4V65 4v66 
4V66 4v67 
4V67 4v68 
4V68 4v69 
4V69 4v6a 
4V6A 4v6c 
4V6C 4v6d 
4V6D 4v6e 
4V6E 4v6f 
4V6F 4v6g 
4V6G 4v6i 
4V6I 4v6k 
4V6K 4v6l 
4V6L 4v6m 
4V6M 4v6n 
4V6N 4v6o 
4V6O 4v6p 
4V6P 4v6q 
4V6Q 4v6r 
4V6R 4v6s 
4V6S 4v6t 
4V6T 4v6u 
4V6U 4v6v 
4V6V 4v6w 
4V6W 4v6x 
4V6X 4v6y 
4V6Y 4v6z 
4V6Z 4v70 
4V70 4v71 
4V71 4v72 
4V72 4v73 
4V73 4v74 
4V74 4v75 
4V75 4v76 
4V76 4v77 
4V77 4v78 
4V78 4v79 
4V79 4v7a 
4V7A 4v7b 
4V7B 4v7c 
4V7C 4v7d 
4V7D 4v7e 
4V7E 4v7h 
4V7H 4v7i 
4V7I 4v7j 
4V7J 4v7k 
4V7K 4v7l 
4V7L 4v7m 
4V7M 4v7p 
4V7P 4v7r 
4V7R 4v7s 
4V7S 4v7t 
4V7T 4v7u 
4V7U 4v7v 
4V7V 4v7w 
4V7W 4v7x 
4V7X 4v7y 
4V7Y 4v7z 
4V7Z 4v83 
4V83 4v84 
4V84 4v85 
4V85 4v87 
4V87 4v88 
4V88 4v89 
4V89 4v8a 
4V8A 4v8b 
4V8B 4v8c 
4V8C 4v8d 
4V8D 4v8e 
4V8E 4v8f 
4V8F 4v8g 
4V8G 4v8h 
4V8H 4v8i 
4V8I 4v8j 
4V8J 4v8n 
4V8N 4v8o 
4V8O 4v8q 
4V8Q 4v8u 
4V8U 4v8x 
4V8X 4v8y 
4V8Y 4v8z 
4V8Z 4v90 
4V90 4v92 
4V92 4v95 
4V95 4v97 
4V97 4v9a 
4V9A 4v9b 
4V9B 4v9c 
4V9C 4v9d 
4V9D 4v9h 
4V9H 4v9i 
4V9I 4v9j 
4V9J 4v9k 
4V9K 4v9l 
4V9L 4v9m 
4V9M 4v9n 
4V9N 4v9o 
4V9O 4v9p 
4V9P 4v9q 
4V9Q 4v9r 
4V9R 4v9s 
4V9S 4w29 
4W29 4w2e 
4W2E 4w2f 
4W2F 4w2g 
4W2G 4w2h 
4W2H 4w2i 
4W2I 4w4g 
4W4G 4wal 
4WAL 4wan 
4WAN 4wf1 
4WF1 4woi 
4WOI 4wpo 
4WPO 4wq1 
4WQ1 4wqf 
4WQF 4wqr 
4WQR 4wqu 
4WQU 4wqy 
4WQY 4wr6 
4WR6 4wra 
4WRA 4wro 
4WRO 4wsd 
4WSD 4wsm 
4WSM 4wt1 
4WT1 4wt8 
4WT8 4wu1 
4WU1 4www 
4WWW 4wzd 
4WZD 4wzo 
4WZO 4x62 
4X62 4x64 
4X64 4x65 
4X65 4x66 
4X66 4xej 
4XEJ 4y4o 
4Y4O 4y4p 
4Y4P 4ybb 
4YBB 4yhh 
4YHH 4ypb 
4YPB 4yy3 
4YY3 4yzv 
4YZV 4z3s 
4Z3S 4z8c 
4Z8C 4zer 
4ZER 4zsn 
4ZSN 5a2q 
5A2Q 5a9z 
5A9Z 5aa0 
5AA0 5afi 
5AFI 5aj0 
5AJ0 5br8 
5BR8 5czp 
5CZP 5d8b 
5D8B 5dat 
5DAT 5dc3 
5DC3 5dfe 
5DFE 5dox 
5DOX 5doy 
5DOY 5e7k 
5E7K 5e81 
5E81 5el3 
5EL3 5el4 
5EL4 5el5 
5EL5 5el6 
5EL6 5el7 
5EL7 5elr 
5ELR 5els 
5ELS 5elt 
5ELT 5emo 
5EMO 5f8k 
5F8K 5fci 
5FCI 5fcj 
5FCJ 5fdu 
5FDU 5fdv 
5FDV 5flx 
5FLX 5hau 
5HAU 5hcp 
5HCP 5hcq 
5HCQ 5hcr 
5HCR 5hd1 
5HD1 5i4l 
5I4L 5ib7 
5IB7 5ib8 
5IB8 5ibb 
5IBB 5imq 
5IMQ 5imr 
5IMR 5iqr 
5IQR 5it7 
5IT7 5it8 
5IT8 5it9 
5IT9 5iwa 
5IWA 5j30 
5J30 5j3c 
5J3C 5j4b 
5J4B 5j4c 
5J4C 5j4d 
5J4D 5j5b 
5J5B 5j7l 
5J7L 5j88 
5J88 5j8a 
5J8A 5j8b 
5J8B 5j91 
5J91 5jb3 
5JB3 5jc9 
5JC9 5jpq 
5JPQ 5jte 
5JTE 5ju8 
5JU8 5juo 
5JUO 5jup 
5JUP 5jus 
5JUS 5jut 
5JUT 5juu 
5JUU 5k0y 
5K0Y 5kcr 
5KCR 5kcs 
5KCS 5kps 
5KPS 5kpv 
5KPV 5kpw 
5KPW 5kpx 
5KPX 5l3p 
5L3P 5lmn 
5LMN 5lmo 
5LMO 5lmp 
5LMP 5lmq 
5LMQ 5lmr 
5LMR 5lms 
5LMS 5lmt 
5LMT 5lmu 
5LMU 5lmv 
5LMV 5lyb 
5LYB 5lza 
5LZA 5lzb 
5LZB 5lzc 
5LZC 5lzd 
5LZD 5lze 
5LZE 5lzf 
5LZF 5lzs 
5LZS 5lzt 
5LZT 5lzu 
5LZU 5lzv 
5LZV 5lzw 
5LZW 5lzx 
5LZX 5lzy 
5LZY 5lzz 
5LZZ 5tga 
5TGA - Links (links to other resources describing this domain)
-
PFAM KH-domain INTERPRO IPR004087

