The domain within your query sequence starts at position 58 and ends at position 174; the E-value for the PA2c domain shown below is 1.75e-15.
All catalytic sites are present in this domain. Check the literature (PubMed 99175087 ) for details.
LLLQFVNSMRCVTGLCPRDFEDYGCACRFEMEGMPVDESDICCFQHRRCYEEAVEMDCLQ DPAKLSADVDCTNKQITCESEDPCERLLCTCDKAAVECLAQSGINSSLNFLDASFCL
PA2cPhospholipase A2 |
|---|
| SMART accession number: | SM00085 |
|---|---|
| Description: | - |
| Interpro abstract (IPR016090): | Proteins containing this domain include eukaryotic phospholipase A2 enzymes (PLA2; EC 3.1.1.4 ), small lipolytic enzymes that releases fatty acids from the second carbon group of glycerol, usually in a metal-dependent reaction, to generate lysophospholipid (LysoPL) and a free fatty acid (FA) [ (PUBMED:11872155) ]. The resulting products are either dietary or used in synthetic pathways for leukotrienes and prostaglandins. Often, arachidonic acid is released as a free fatty acid and acts as second messenger in signaling networks [ (PUBMED:10331081) ]. These enzymes enable the of fatty acids and lysophospholipid by hydrolysing the 2-ester bond of 1,2-diacyl-3-sn-phosphoglycerides. In eukaryotes, PLA2 plays a pivotal role in the biosynthesis of prostaglandin and other mediators of inflammation. These enzymes are either secreted or cytosolic; the latter are either Ca dependent or Ca independent. Secreted PLA2s have also been found to specifically bind to a variety of soluble and membrane proteins in mammals, including receptors [ (PUBMED:11293116) ]. As a toxin, PLA2 is a potent presynaptic neurotoxin which blocks nerve terminals by binding to the nerve membrane and hydrolyzing stable membrane lipids [ (PUBMED:11212293) ]. The products of the hydrolysis (LysoPL and FA) cannot form bilayers leading to a change in membrane conformation and ultimately to a block in the release of neurotransmitters [ (PUBMED:16805767) (PUBMED:10838563) (PUBMED:16339444) ]. The phospholipase domain adopts an alpha-helical secondary structure, consisting of five alpha-helices and two helical segments. PLA2 may form dimers or oligomers [ (PUBMED:11080675) (PUBMED:11897785) (PUBMED:12161451) ]. |
| GO process: | arachidonic acid secretion (GO:0050482), phospholipid metabolic process (GO:0006644) |
| GO function: | phospholipase A2 activity (GO:0004623) |
| Family alignment: |
There are 5804 PA2c domains in 5499 proteins in SMART's nrdb database.
Click on the following links for more information.
- Evolution (species in which this domain is found)
-
Taxonomic distribution of proteins containing PA2c domain.
This tree includes only several representative species. The complete taxonomic breakdown of all proteins with PA2c domain is also avaliable.
Click on the protein counts, or double click on taxonomic names to display all proteins containing PA2c domain in the selected taxonomic class.
- Literature (relevant references for this domain)
-
Primary literature is listed below; Automatically-derived, secondary literature is also avaliable.
- Cirino G
- Multiple controls in inflammation. Extracellular and intracellular phospholipase A2, inducible and constitutive cyclooxygenase, and inducible nitric oxide synthase.
- Biochem Pharmacol. 1998; 55: 105-11
- Display abstract
Inflammation occurs as a defensive response to invasion of the host by foreign material, often of microbial nature. This response is normally a localized protective response that at the microscopic level involves a complex series of events including dilatation of arterioles, venules, and capillaries with increased vascular permeability, exudation of fluids including plasma proteins, and leukocyte migration into the inflammatory area. Since disease characterized by inflammation is an important cause of morbidity and mortality in humans, the processes involved in the host defense in inflammation have been and continue to be the object of several experimental studies. The role of several mediators such as histamine, serotonin, bradykinin, prostaglandins, and, more recently, cytokines and nitric oxide has been evaluated, and a contribution for each one of these mediators has been proposed. With the development of powerful molecular biology tools, it has become possible to study enzymes involved in this complex phenomenon by measuring the expression or evaluating the signaling pathways following a specific stimulus. These techniques have generated a proliferation of studies on the role of several enzymes and cytokines in inflammation. Most of these studies have been conducted in vitro on cell lines, and not many of the results have been confirmed by in vivo studies. This commentary does not pretend to analyze all of the studies and their possible in congruences, but endeavors to provoke in the reader a critical review of dogmas and current beliefs that most of the time are built on unilateral interpretation of the data.
- Davidson FF, Dennis EA
- Evolutionary relationships and implications for the regulation of phospholipase A2 from snake venom to human secreted forms.
- J Mol Evol. 1990; 31: 228-38
- Display abstract
The amino acid sequences of 40 secreted phospholipase A2's (PLA2) were aligned and a phylogenetic tree derived that has three main branches corresponding to elapid (group I), viperid (group II), and insect venom types of PLA2. The human pancreatic and recently determined nonpancreatic sequences in the comparison align with the elapid and viperid categories, respectively, indicating that at least two PLA2 genes existed in the vertebrate line before the divergence of reptiles and mammals about 200-300 million years ago. This allows resolution for the first time of major genetic events in the evolution of current PLA2's and the relationship of human PLA2's to those of snake venom, many of which are potent toxins. Implications for possible mechanisms of regulation of mammalian intra- and extracellular PLA2's are discussed, as well as issues relating to the search for the controlling enzymes in arachidonic acid release, prostaglandin generation, and signal transduction.
- Gomez F et al.
- Purification and characterization of five variants of phospholipase A2 and complete primary structure of the main phospholipase A2 variant in Heloderma suspectum (Gila monster) venom.
- Eur J Biochem. 1989; 186: 23-33
- Display abstract
1. Five increasingly anionic variants (Pa1-Pa5) of Ca2+-dependent phospholipase A2 were purified to homogeneity from the venom of the lizard Heloderma suspectum (Gila monster). The purification procedure was based on semi-preparative reverse-phase HPLC followed by anion-exchange HPLC and analytical reverse-phase HPLC. 2. Their Mr were 17,000-18,000, as deduced by SDS/PAGE. Specific activities tested by the capacity to hydrolyze phosphatidylcholines at pH 8.5 decreased as follows: Pa3 greater than Pa5 greater than Pa4 greater than Pa1 greater than Pa2. These activities showed the same optimum pH (9.0), were mainly of the phospholipase A2 type and were lost upon p-bromophenacyl bromide treatment. 3. All five phospholipases efficiently stimulated amylase release from dispersed rat pancreatic acini at pH 7.4, their potency decreasing as follows: Pa2 greater than Pa1 approximately equal to Pa4 greater than Pa3 approximately equal to Pa5. No deleterious effect was apparent based on the lack of lactate dehydrogenase release. 4. The five variants, Pa1-Pa5, differed significantly in amino acid composition and this, together with distinct antigenic properties of Pa2 and Pa5, establishes the subheterogeneity of this new type of phospholipase A2, despite the fact that the N-terminal amino acid sequence (31 residues) of Pa1-Pa5 was exactly the same. 5. The full sequence of the major variant, Pa5, showed that this 142-amino-acid protein exhibited greater similarity to the bee venom enzyme than to any class I or class II secretory phospholipase A2 from snake venom and mammalian pancreas. While Pa5 displayed the highly conserved region between Asp30 and Cys39 (the essential active site of all phospholipases A2), its salient original points included 10 half-cystine residues only, an incomplete N-terminal sequence, large changes in the putative calcium loop, several alterations after the active site and a C-terminal extension never seen in other phospholipases A2, with the only exception being bee venom.
- Dijkstra BW, Renetseder R, Kalk KH, Hol WG, Drenth J
- Structure of porcine pancreatic phospholipase A2 at 2.6 A resolution and comparison with bovine phospholipase A2.
- J Mol Biol. 1983; 168: 163-79
- Display abstract
The previously published three-dimensional structure of porcine pancreatic prophospholipase A2 at 3 A resolution was found to be incompatible with the structures of bovine phospholipase A2 and bovine prophospholipase A2. This was unexpected because of the very homologous amino acid sequences of these enzymes. Therefore, the crystal structure of the porcine enzyme was redetermined using molecular replacement methods with bovine phospholipase as the parent model. The structure was crystallographically refined at 2.6 A resolution by fast Fourier transform and restrained least-squares procedures to an R-factor of 0.241. The crystals appeared to contain phospholipase A2 and not prophospholipase A2. Apparently the protein is slowly converted under the crystallization conditions employed. Our investigation shows that, in contrast to the previous report, the three-dimensional structure of porcine phospholipase A2 is very similar to that of bovine phospholipase A2, including the active site. Smaller differences were observed in some residues involved in the binding of aggregated substrates. However, an appreciable conformational difference is in the loop 59 to 70, where a single substitution at position 63 (bovine Val leads to porcine Phe) causes a complete rearrangement of the peptide chain. In addition to the calcium ion in the active site, a second calcium ion is present in the crystals; this is located on a crystallographic 2-fold axis and stabilizes the interaction between two neighbouring molecules.
- Metabolism (metabolic pathways involving proteins which contain this domain)
-
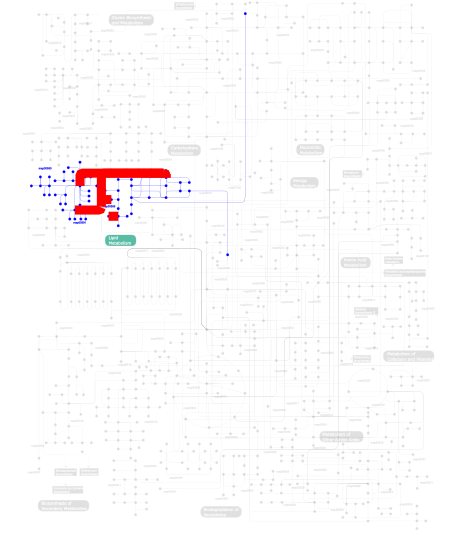
Click the image to view the interactive version of the map in iPath% proteins involved KEGG pathway ID Description 10.00 map04664 Fc epsilon RI signaling pathway 10.00  map00590
map00590Arachidonic acid metabolism 10.00  map00565
map00565Ether lipid metabolism 10.00  map00564
map00564Glycerophospholipid metabolism 10.00 map04370 VEGF signaling pathway 10.00 map04010 MAPK signaling pathway 10.00 map04912 GnRH signaling pathway 10.00 map00592 alpha-Linolenic acid metabolism 10.00 map04730 Long-term depression 10.00  map00591
map00591Linoleic acid metabolism This information is based on mapping of SMART genomic protein database to KEGG orthologous groups. Percentage points are related to the number of proteins with PA2c domain which could be assigned to a KEGG orthologous group, and not all proteins containing PA2c domain. Please note that proteins can be included in multiple pathways, ie. the numbers above will not always add up to 100%.
- Structure (3D structures containing this domain)
3D Structures of PA2c domains in PDB
PDB code Main view Title 1a2a 
AGKISTROTOXIN, A PHOSPHOLIPASE A2-TYPE PRESYNAPTIC NEUROTOXIN FROM AGKISTRODON HALYS PALLAS 1a3d 
PHOSPHOLIPASE A2 (PLA2) FROM NAJA NAJA VENOM 1a3f 
PHOSPHOLIPASE A2 (PLA2) FROM NAJA NAJA VENOM 1ae7 
NOTEXIN, A PRESYNAPTIC NEUROTOXIC PHOSPHOLIPASE A2 1aok 
VIPOXIN COMPLEX 1ayp 
A PROBE MOLECULE COMPOSED OF SEVENTEEN PERCENT OF TOTAL DIFFRACTING MATTER GIVES CORRECT SOLUTIONS IN MOLECULAR REPLACEMENT 1b4w 
BASIC PHOSPHOLIPASE A2 FROM AGKISTRODON HALYS PALLAS-IMPLICATIONS FOR ITS ASSOCIATION AND ANTICOAGULANT ACTIVITIES BY X-RAY CRYSTALLOGRAPHY 1bbc 
STRUCTURE OF RECOMBINANT HUMAN RHEUMATOID ARTHRITIC SYNOVIAL FLUID PHOSPHOLIPASE A2 AT 2.2 ANGSTROMS RESOLUTION 1bjj 
AGKISTRODOTOXIN, A PHOSPHOLIPASE A2-TYPE PRESYNAPTIC NEUROTOXIN FROM AGKISTRODON HALYS PALLAS 1bk9 
PHOSPHOLIPASE A2 MODIFIED BY PBPB 1bp2 
STRUCTURE OF BOVINE PANCREATIC PHOSPHOLIPASE A2 AT 1.7 ANGSTROMS RESOLUTION 1bpq 
PHOSPHOLIPASE A2 ENGINEERING. X-RAY STRUCTURAL AND FUNCTIONAL EVIDENCE FOR THE INTERACTION OF LYSINE-56 WITH SUBSTRATES 1bun 
STRUCTURE OF BETA2-BUNGAROTOXIN: POTASSIUM CHANNEL BINDING BY KUNITZ MODULES AND TARGETED PHOSPHOLIPASE ACTION 1bvm 
SOLUTION NMR STRUCTURE OF BOVINE PANCREATIC PHOSPHOLIPASE A2, 20 STRUCTURES 1c1j 
STRUCTURE OF CADMIUM-SUBSTITUTED PHOSPHOLIPASE A2 FROM AGKISTRONDON HALYS PALLAS AT 2.8 ANGSTROMS RESOLUTION 1c74 
Structure of the double mutant (K53,56M) of phospholipase A2 1ceh 
STRUCTURE AND FUNCTION OF THE CATALYTIC SITE MUTANT ASP99ASN OF PHOSPHOLIPASE A2: ABSENCE OF CONSERVED STRUCTURAL WATER 1cl5 
CRYSTAL STRUCTURE OF PHOSPHOLIPASE A2 FROM DABOIA RUSSELLI PULCHELLA 1clp 
CRYSTAL STRUCTURE OF A CALCIUM-INDEPENDENT PHOSPHOLIPASELIKE MYOTOXIC PROTEIN FROM BOTHROPS ASPER VENOM 1db4 
HUMAN S-PLA2 IN COMPLEX WITH INDOLE 8 1db5 
HUMAN S-PLA2 IN COMPLEX WITH INDOLE 6 1dcy 
CRYSTAL STRUCTURE OF HUMAN S-PLA2 IN COMPLEX WITH INDOLE 3 ACTIVE SITE INHIBITOR 1dpy 
THREE-DIMENSIONAL STRUCTURE OF A NOVEL PHOSPHOLIPASE A2 FROM INDIAN COMMON KRAIT AT 2.45 A RESOLUTION 1fb2 
STRUCTURE OF PHOSPHOLIPASE A2 FROM DABOIA RUSSELLI PULCHELLA AT 1.95 1fdk 
CARBOXYLIC ESTER HYDROLASE (PLA2-MJ33 INHIBITOR COMPLEX) 1fe5 
SEQUENCE AND CRYSTAL STRUCTURE OF A BASIC PHOSPHOLIPASE A2 FROM COMMON KRAIT (BUNGARUS CAERULEUS) AT 2.4 RESOLUTION: IDENTIFICATION AND CHARACTERIZATION OF ITS PHARMACOLOGICAL SITES. 1fv0 
FIRST STRUCTURAL EVIDENCE OF THE INHIBITION OF PHOSPHOLIPASE A2 BY ARISTOLOCHIC ACID: CRYSTAL STRUCTURE OF A COMPLEX FORMED BETWEEN PHOSPHOLIPASE A2 AND ARISTOLOCHIC ACID 1fx9 
CARBOXYLIC ESTER HYDROLASE COMPLEX (DIMERIC PLA2 + MJ33 INHIBITOR + SULPHATE IONS) 1fxf 
CARBOXYLIC ESTER HYDROLASE COMPLEX (DIMERIC PLA2 + MJ33 INHIBITOR + PHOSPHATE IONS) 1g0z 
SPECIFIC MUTATIONS IN KRAIT PLA2 LEAD TO DIMERIZATION OF PROTEIN MOLECULES: CRYSTAL STRUCTURE OF KRAIT PLA2 AT 2.1 RESOLUTION 1g2x 
Sequence induced trimerization of krait PLA2: crystal structure of the trimeric form of krait PLA2 1g4i 
Crystal structure of the bovine pancreatic phospholipase A2 at 0.97A 1gh4 
Structure of the triple mutant (K56M, K120M, K121M) of phospholipase A2 1gmz 
Crystal structure of the D49 phospholipase A2 piratoxin III from Bothrops pirajai. 1god 
MONOMERIC LYS-49 PHOSPHOLIPASE A2 HOMOLOGUE ISOLATED FROM THE VENOM OF CERROPHIDION (BOTHROPS) GODMANI 1gp7 
Acidic Phospholipase A2 from venom of Ophiophagus Hannah 1hn4 
PROPHOSPHOLIPASE A2 DIMER COMPLEXED WITH MJ33, SULFATE, AND CALCIUM 1ijl 
Crystal structure of acidic phospholipase A2 from deinagkistrodon acutus 1irb 
CARBOXYLIC ESTER HYDROLASE 1j1a 
PANCREATIC SECRETORY PHOSPHOLIPASE A2 (IIa) WITH ANTI-INFLAMMATORY ACTIVITY 1jia 
STRUCTURE OF A BASIC PHOSPHOLIPASE A2 FROM AGKISTRODON HALYS PALLAS AT 2.13A RESOLUTION 1jlt 
Vipoxin Complex 1jq8 
Design of specific inhibitors of phospholipase A2: Crystal structure of a complex formed between phospholipase A2 from Daboia russelli pulchella and a designed pentapeptide Leu-Ala-Ile-Tyr-Ser at 2.0 resolution 1jq9 
Crystal structure of a complex formed between phospholipase A2 from Daboia russelli pulchella and a designed pentapeptide Phe-Leu-Ser-Tyr-Lys at 1.8 resolution 1kpm 
First Structural Evidence of a Specific Inhibition of Phospholipase A2 by Vitamin E and its Implications in Inflammation: Crystal Structure of the Complex Formed between Phospholipase A2 and Vitamin E at 1.8 A Resolution. 1kqu 
Human phospholipase A2 complexed with a substrate anologue 1kvo 
HUMAN PHOSPHOLIPASE A2 COMPLEXED WITH A HIGHLY POTENT SUBSTRATE ANOLOGUE 1kvw 
CARBOXYLIC ESTER HYDROLASE, SINGLE MUTANT H48Q OF BOVINE PANCREATIC PLA2 ENZYME 1kvx 
CARBOXYLIC ESTER HYDROLASE, SINGLE MUTANT D99A OF BOVINE PANCREATIC PLA2, 1.9 A ORTHORHOMBIC FORM 1kvy 
CARBOXYLIC ESTER HYDROLASE, SINGLE MUTANT D49E COORDINATED TO CALCIUM 1l8s 
CARBOXYLIC ESTER HYDROLASE COMPLEX (DIMERIC PLA2 + LPC-ether + ACETATE + PHOSPHATE IONS) 1le6 
CARBOXYLIC ESTER HYDROLASE, P 1 21 1 SPACE GROUP 1le7 
CARBOXYLIC ESTER HYDROLASE, C 2 2 21 space group 1ln8 
Crystal Structure of a New Isoform of Phospholipase A2 from Naja naja sagittifera at 1.6 A Resolution 1m8r 
Crystal Structures of Cadmium-binding Acidic Phospholipase A2 from the Venom of Agkistrodon halys pallas at 1.9 Resolution (crystal grown at pH 7.4) 1m8s 
Crystal Structures of Cadmium-binding Acidic Phospholipase A2 from the Venom of Agkistrodon halys pallas at 1.9 Resolution (crystal grown at pH 5.9) 1m8t 
Structure of an acidic Phospholipase A2 from the venom of Ophiophagus hannah at 2.1 resolution from a hemihedrally twinned crystal form 1mc2 
monomeric LYS-49 phospholipase A2 homologue purified from AG 1mf4 
Structure-based design of potent and selective inhibitors of phospholipase A2: Crystal structure of the complex formed between phosholipase A2 from Naja Naja sagittifera and a designed peptide inhibitor at 1.9 A resolution 1mg6 
The Crystal Structure of a K49 PLA2 from the Snake Venom of Agkistrodon acutus 1mh2 
Crystal Structure of a Zinc Containing Dimer of Phospholipase A2 from the Venom of Indian Cobra (Naja Naja Sagittifera) 1mh7 
Crystal Structure of a Calcium-Free Isoform of Phospholipase A2 from Naja naja sagittifera at 2.0 A Resolution 1mh8 
Crystal Structure of a Phopholipase A2 Monomer with Isoleucine at Second Position 1mks 
CARBOXYLIC ESTER HYDROLASE, TRIGONAL FORM OF THE TRIPLE MUTANT 1mkt 
CARBOXYLIC ESTER HYDROLASE, 1.72 ANGSTROM TRIGONAL FORM OF THE BOVINE RECOMBINANT PLA2 ENZYME 1mku 
CARBOXYLIC ESTER HYDROLASE, ORTHORHOMBIC FORM OF THE TRIPLE MUTANT 1mkv 
CARBOXYLIC ESTER HYDROLASE COMPLEX (PLA2 + TRANSITION STATE ANALOG COMPLEX) 1n28 
Crystal structure of the H48Q mutant of human group IIA phospholipase A2 1n29 
Crystal structure of the N1A mutant of human group IIA phospholipase A2 1o2e 
Structure of the triple mutant (K53,56,120M) + Anisic acid complex of phospholipase A2 1o3w 
Structure of the inhibitor free triple mutant (K53,56,120M) of phospholipase A2 1oqs 
Crystal Structure of RV4/RV7 Complex 1ows 
Crystal structure of a C49 Phospholipase A2 from Indian cobra reveals carbohydrate binding in the hydrophobic channel 1oxl 
INHIBITION OF PHOSPHOLIPASE A2 (PLA2) BY (2-CARBAMOYLMETHYL-5-PROPYL-OCTAHYDRO-INDOL-7-YL)-ACETIC ACID (INDOLE): CRYSTAL STRUCTURE OF THE COMPLEX FORMED BETWEEN PLA2 FROM RUSSELL'S VIPER AND INDOLE AT 1.8 RESOLUTION 1oxr 
Aspirin induces its Anti-inflammatory effects through its specific binding to Phospholipase A2: Crystal structure of the complex formed between Phospholipase A2 and Aspirin at 1.9A resolution 1oyf 
Crystal Structure of Russelles viper (Daboia russellii pulchella) phospholipase A2 in a complex with venom 6-methyl heptanol 1oz6 
X-ray structure of acidic phospholipase A2 from Indian saw-scaled viper (Echis carinatus) with a potent platelet aggregation inhibitory activity 1ozy 
Crystal Structure of Phospholipase A2 (MIPLA3) From Micropechis Ikaheka 1p2p 
STRUCTURE OF PORCINE PANCREATIC PHOSPHOLIPASE A2 AT 2.6 ANGSTROMS RESOLUTION AND COMPARISON WITH BOVINE PHOSPHOLIPASE A2 1p7o 
Crystal structure of phospholipase A2 (MIPLA4) from Micropechis ikaheka 1pa0 
CRYSTAL STRUCTURE OF BNSP-7, A LYS49-PHOSPHOLIPASE A2 1pc9 
Crystal Structure of BnSP-6, a Lys49-Phospholipase A2 1pir 
SOLUTION STRUCTURE OF PORCINE PANCREATIC PHOSPHOLIPASE A2 1pis 
SOLUTION STRUCTURE OF PORCINE PANCREATIC PHOSPHOLIPASE A2 1po8 
Crystal structure of a complex formed between krait venom phospholipase A2 and heptanoic acid at 2.7 A resolution. 1poa 
INTERFACIAL CATALYSIS: THE MECHANISM OF PHOSPHOLIPASE A2 1pob 
CRYSTAL STRUCTURE OF COBRA-VENOM PHOSPHOLIPASE A2 IN A COMPLEX WITH A TRANSITION-STATE ANALOGUE 1poc 
CRYSTAL STRUCTURE OF BEE-VENOM PHOSPHOLIPASE A2 IN A COMPLEX WITH A TRANSITION-STATE ANALOGUE 1pod 
STRUCTURES OF FREE AND INHIBITED HUMAN SECRETORY PHOSPHOLIPASE A2 FROM INFLAMMATORY EXUDATE 1poe 
STRUCTURES OF FREE AND INHIBITED HUMAN SECRETORY PHOSPHOLIPASE A2 FROM INFLAMMATORY EXUDATE 1pp2 
THE REFINED CRYSTAL STRUCTURE OF DIMERIC PHOSPHOLIPASE A2 AT 2.5 ANGSTROMS. ACCESS TO A SHIELDED CATALYTIC CENTER 1ppa 
THE CRYSTAL STRUCTURE OF A LYSINE 49 PHOSPHOLIPASE A2 FROM THE VENOM OF THE COTTONMOUTH SNAKE AT 2.0 ANGSTROMS RESOLUTION 1psh 
CRYSTAL STRUCTURE OF PHOSPHOLIPASE A2 FROM INDIAN COBRA REVEALS A TRIMERIC ASSOCIATION 1psj 
ACIDIC PHOSPHOLIPASE A2 FROM AGKISTRODON HALYS PALLAS 1pwo 
Crystal Structure of Phospholipase A2 (MIPLA2) from Micropechis Ikaheka 1q5t 
Gln48 PLA2 separated from Vipoxin from the venom of Vipera ammodytes meridionalis. 1q6v 
First crystal structure of a C49 monomer PLA2 from the venom of Daboia russelli pulchella at 1.8 A resolution 1q7a 
Crystal structure of the complex formed between russell's viper phospholipase A2 and an antiinflammatory agent oxyphenbutazone at 1.6A resolution 1qll 
Piratoxin-II (Prtx-II) - a K49 PLA2 from Bothrops pirajai 1rgb 
Phospholipase A2 from Vipera ammodytes meridionalis 1s6b 
X-ray Crystal Structure of a Complex Formed Between Two Homologous Isoforms of Phospholipase A2 from Naja naja sagittifera: Principle of Molecular Association and Inactivation 1s8g 
Crystal structure of Lys49-Phospholipase A2 from Agkistrodon contortrix laticinctus, fatty acid bound form 1s8h 
Crystal structure of Lys49-Phospholipase A2 from Agkistrodon contortrix laticinctus, first fatty acid free form 1s8i 
Crystal structure of Lys49-Phospholipase A2 from Agkistrodon contortrix laticinctus, second fatty acid free form 1sfv 
PORCINE PANCREAS PHOSPHOLIPASE A2, NMR, MINIMIZED AVERAGE STRUCTURE 1sfw 
PORCINE PANCREAS PHOSPHOLIPASE A2, NMR, 18 STRUCTURES 1skg 
Structure-based rational drug design: Crystal structure of the complex formed between Phospholipase A2 and a pentapeptide Val-Ala-Phe-Arg-Ser 1sqz 
Design of specific inhibitors of Phopholipase A2: Crystal structure of the complex formed between Group II Phopholipase A2 and a designed peptide Dehydro-Ile-Ala-Arg-Ser at 1.2A resolution 1sv3 
Structure of the complex formed between Phospholipase A2 and 4-methoxybenzoic acid at 1.3A resolution. 1sv9 
Crystal structure of the complex formed between groupII phospholipase A2 and anti-inflammatory agent 2-[(2,6-Dichlorophenyl)amino] benzeneacetic acid at 2.7A resolution 1sxk 
Crystal Structure of a complex formed between phospholipase A2 and a non-specific anti-inflammatory amino salicylic acid at 1.2 A resolution 1sz8 
Crystal Structure of an Acidic Phospholipase A2 from Naja Naja Sagittifera at 1.5 A resolution 1t37 
Design of specific inhibitors of phospholipase A2: Crystal structure of the complex formed between group I phospholipase A2 and a designed pentapeptide Leu-Ala-Ile-Tyr-Ser at 2.6A resolution 1tc8 
Crystal structure of Krait-venom phospholipase A2 in a complex with a natural fatty acid tridecanoic acid 1td7 
Interactions of a specific non-steroidal anti-inflammatory drug (NSAID) with group I phospholipase A2 (PLA2): Crystal structure of the complex formed between PLA2 and niflumic acid at 2.5 A resolution 1tdv 
Non-specific binding to phospholipase A2:Crystal structure of the complex of PLA2 with a designed peptide Tyr-Trp-Ala-Ala-Ala-Ala at 1.7A resolution 1tg1 
Crystal Structure of the complex formed between russells viper phospholipase A2 and a designed peptide inhibitor PHQ-Leu-Val-Arg-Tyr at 1.2A resolution 1tg4 
Design of specific inhibitors of groupII phospholipase A2(PLA2): Crystal structure of the complex formed between russells viper PLA2 and designed peptide Phe-Leu-Ala-Tyr-Lys at 1.7A resolution 1tgm 
Crystal structure of a complex formed between group II phospholipase A2 and aspirin at 1.86 A resolution 1th6 
Crystal structure of phospholipase A2 in complex with atropine at 1.23A resolution 1tj9 
Structure of the complexed formed between group II phospholipase A2 and a rationally designed tetra peptide,Val-Ala-Arg-Ser at 1.1A resolution 1tjk 
Crystal structure of the complex formed between group II phospholipase A2 with a designed pentapeptide, Phe- Leu- Ser- Thr- Lys at 1.2 A resolution 1tk4 
Crystal structure of russells viper phospholipase A2 in complex with a specifically designed tetrapeptide Ala-Ile-Arg-Ser at 1.1 A resolution 1tp2 
Crystal structure of the complex of group II phospholipaseA2 dimer with a fatty acid tridecanoic acid at 2.4 A resolution 1u4j 
Crystal structure of a carbohydrate induced dimer of group I phospholipase A2 from Bungarus caeruleus at 2.1 A resolution 1u73 
Crystal structure of a Dimeric Acidic Platelet Aggregation Inhibitor and Hypotensive Phospholipase A2 from Bothrops jararacussu 1umv 
Crystal structure of an acidic, non-myotoxic phospholipase A2 from the venom of Bothrops jararacussu 1une 
CARBOXYLIC ESTER HYDROLASE, 1.5 ANGSTROM ORTHORHOMBIC FORM OF THE BOVINE RECOMBINANT PLA2 1vap 
THE MONOMERIC ASP49 SECRETORY PHOSPHOLIPASE A2 FROM THE VENOM OF AGKISTRIDON PISCIVORUS PISCIVORUS 1vip 
ANTICOAGULANT CLASS II PHOSPHOLIPASE A2 FROM THE VENOM OF VIPERA RUSSELLI RUSSELLI 1vkq 
A re-determination of the structure of the triple mutant (K53,56,120M) of phospholipase A2 at 1.6A resolution using sulphur-SAS at 1.54A wavelength 1vl9 
Atomic resolution (0.97A) structure of the triple mutant (K53,56,121M) of bovine pancreatic phospholipase A2 1vpi 
PHOSPHOLIPASE A2 INHIBITOR FROM VIPOXIN 1xxs 
Structural insights for fatty acid binding in a Lys49 phospholipase A2: crystal structure of myotoxin II from Bothrops moojeni complexed with stearic acid 1xxw 
Structure of zinc induced heterodimer of two calcium free isoforms of phospholipase A2 from Naja naja sagittifera at 2.7A resolution 1y38 
Crystal structure of the complex formed between phospholipase A2 dimer and glycerophosphate at 2.4 A resolution 1y4l 
Crystal structure of Bothrops asper myotoxin II complexed with the anti-trypanosomal drug suramin 1y6o 
Crystal structure of disulfide engineered porcine pancreatic phospholipase A2 to group-X isozyme in complex with inhibitor MJ33 and phosphate ions 1y6p 
Crystal structure of disulfide engineered porcine pancratic phospholipase a2 to group-x isozyme 1y75 
A new form of catalytically inactive phospholipase A2 with an unusual disulphide bridge Cys 32- Cys 49 reveals recognition for N-acetylglucosmine 1yxh 
Crystal structure of a novel phospholipase A2 from Naja naja sagittifera with a strong anticoagulant activity 1yxl 
Crystal structure of a novel phospholipase A2 from Naja naja sagittifera at 1.5 A resolution 1z76 
Crystal structure of an acidic phospholipase A2 (BthA-I) from Bothrops jararacussu venom complexed with p-bromophenacyl bromide 1zl7 
Crystal structure of catalytically-active phospholipase A2 with bound calcium 1zlb 
Crystal structure of catalytically-active phospholipase A2 in the absence of calcium 1zm6 
Crystal structure of the complex formed beween a group I phospholipase A2 and designed penta peptide Leu-Ala-Ile-Tyr-Ser at 2.6A resolution 1zr8 
Crystal Structure of the complex formed between group II phospholipase A2 and a plant alkaloid ajmaline at 2.0A resolution 1zwp 
The atomic resolution Crystal structure of the Phospholipase A2 (PLA2) complex with Nimesulide reveals its weaker binding to PLA2 1zyx 
Crystal structure of the complex of a group IIA phospholipase A2 with a synthetic anti-inflammatory agent licofelone at 1.9A resolution 2aoz 
Crystal structure of the myotoxin-II from Atropoides nummifer venom 2arm 
Crystal Structure of the Complex of Phospholipase A2 with a natural compound atropine at 1.2 A resolution 2azy 
Crystal Structure of Porcine Pancreatic Phospholipase A2 in Complex with Cholate 2azz 
Crystal Structure of Porcine Pancreatic Phospholipase A2 in Complex with Taurocholate 2b00 
Crystal Structure of Porcine Pancreatic Phospholipase A2 in Complex with Glycocholate 2b01 
Crystal Structure of Porcine Pancreatic Phospholipase A2 in Complex with Taurochenodeoxycholate 2b03 
Crystal Structure of Porcine Pancreatic Phospholipase A2 in Complex with Taurochenodeoxycholate 2b04 
Crystal Structure of Porcine Pancreatic Phospholipase A2 in Complex with Glycochenodeoxycholate 2b17 
Specific binding of non-steroidal anti-inflammatory drugs (NSAIDs) to phospholipase A2: Crystal structure of the complex formed between phospholipase A2 and diclofenac at 2.7 A resolution: 2b96 
Third Calcium ion found in an inhibitor bound phospholipase A2 2bax 
Atomic Resolution Structure of the Double Mutant (K53,56M) of Bovine Pancreatic Phospholipase A2 2bch 
A possible of Second calcium ion in interfacial binding: Atomic and Medium resolution crystal structures of the quadruple mutant of phospholipase A2 2bd1 
A possible role of the second calcium ion in interfacial binding: Atomic and medium resolution crystal structures of the quadruple mutant of phospholipase A2 2bp2 
THE STRUCTURE OF BOVINE PANCREATIC PROPHOSPHOLIPASE A2 AT 3.0 ANGSTROMS RESOLUTION 2bpp 
PHOSPHOLIPASE A2 ENGINEERING. X-RAY STRUCTURAL AND FUNCTIONAL EVIDENCE FOR THE INTERACTION OF LYSINE-56 WITH SUBSTRATES 2do2 
Design of specific inhibitors of phospholipase A2: Crystal structure of the complex formed between a group II Cys 49 phospholipase A2 and a designed pentapeptide Ala-Leu-Ala-Ser-Lys at 2.6A resolution 2dpz 
Structure of the complex of phospholipase A2 with N-(4-hydroxyphenyl)- acetamide at 2.1 A resolution 2fnx 
Design of Specific Peptide Inhibitors of Phospholipase A2 (PLA2): Crystal Structure of the Complex of PLA2 with a Highly Potent Peptide Val-Ile-Ala-Lys at 2.7A Resolution 2g58 
Crystal structure of a complex of phospholipase A2 with a designed peptide inhibitor Dehydro-Ile-Ala-Arg-Ser at 0.98 A resolution 2gns 
Design of specific peptide inhibitors of phospholipase A2: Crystal structure of the complex formed between a group II phospholipase A2 and a designed pentapeptide Ala- Leu- Val- Tyr- Lys at 2.3 A resolution 2h4c 
Structure of Daboiatoxin (heterodimeric PLA2 venom) 2h8i 
Crystal Structure of the Bothropstoxin-I complexed with polyethylene glycol 2i0u 
Crystal structures of phospholipases A2 from Vipera nikolskii venom revealing Triton X-100 bound in hydrophobic channel 2not 
NOTECHIS II-5, NEUROTOXIC PHOSPHOLIPASE A2 FROM NOTECHIS SCUTATUS SCUTATUS 2o1n 
Crystal structure of a complex of phospholipase A2 with a peptide Ala-Ile-Ala-Ser at 2.8 A resolution 2ok9 
PrTX-I-BPB 2oli 
Crystal structure of the complex formed between a group II phospholipase A2 and an indole derivative at 2.2 A resolution 2oqd 
Crystal Structure of BthTX-II 2osh 
crystal structure of Natratoxin, a snake sPLA2 that blocks A-type K+ channel 2osn 
An alternate description of a crystal structure of phospholipase A2 from Bungarus caeruleus 2otf 
Crystal structure of the complex formed between phospholipase A2 and atenolol at 1.95 A resolution 2oth 
Crystal structure of a ternary complex of phospholipase A2 with indomethacin and nimesulide at 2.9 A resolution 2oub 
Crystal structure of the complex formed between phospholipase A2 and atenolol at 2.75 A resolution 2oyf 
Crystal Structure of the complex of phospholipase A2 with indole acetic acid at 1.2 A resolution 2pb8 
Crystal structure of the complex formed between phospholipase A2 and peptide Ala-Val-Tyr-Ser at 2.0 A resolution 2ph4 
Crystal structure of a novel Arg49 phospholipase A2 homologue from Zhaoermia mangshanensis venom 2phi 
A LARGE CONFORMATIONAL CHANGE IS FOUND IN THE CRYSTAL STRUCTURE OF THE PORCINE PANCREATIC PHOSPHOLIPASE A2 POINT MUTANT F63V 2pmj 
Crystal structure of the complex formed between phospholipase A2 and 1, 2 benzopyrone at 2.4 A resolution 2pvt 
Crystal structure of a new isoform of phospholipase A2 from russells viper at 2.1 A resolution 2pws 
Crystal structure of the complex formed between phospholipase A2 and 2-(4-isobutyl-phenyl)-propionic acid at 2.2 A resolution 2pyc 
Crystal structure of a monomeric phospholipase A2 from Russell's viper at 1.5A resolution 2q1p 
Crystal Structure of Phospholipase A2 complex with propanol at 1.5 A resolution 2q2j 
Crystal structure of PrTX-I, a PLA2 homolog from Bothrops pirajai 2qhd 
Crystal structure of ecarpholin S (ser49-PLA2) complexed with fatty acid 2qhe 
Crystal structure of Ser49-PLA2 (ecarpholin S) from Echis carinatus sochureki snake venom 2qhw 
Crystal structure of a complex of phospholipase A2 with a gramine derivative at 2.2 resolution 2qog 
Crotoxin B, the basic PLA2 from Crotalus durissus terrificus. 2qu9 
Crystal structure of the complex of group II phospholipase A2 with Eugenol 2que 
Saturation of substrate-binding site using two natural ligands: Crystal structure of a ternary complex of phospholipase A2 with anisic acid and ajmaline at 2.25 A resolution 2qvd 
Identification of a potent anti-inflammatory agent from the natural extract of plant Cardiospermun helicacabum: Crystal structure of the complex of phospholipase A2 with Benzo(g)-1,3-benzodioxolo(5,6-a)quinolizinium, 5,6-dihydro-9,10-dimethoxy at 1.93 A resolution 2rd4 
Design of specific inhibitors of Phospholipase A2: Crystal structure of the complex of phospholipase A2 with pentapeptide Leu-Val-Phe-Phe-Ala at 2.9 A resolution 2wg7 
Structure of Oryza Sativa (Rice) PLA2 2wg8 
Structure of Oryza Sativa (Rice) PLA2, orthorhombic crystal form 2wg9 
Structure of Oryza Sativa (Rice) PLA2, complex with octanoic acid 2wq5 
Non-antibiotic properties of tetracyclines: structural basis for inhibition of secretory phospholipase A2. 2zbh 
Crystal structure of the complex of phospholipase A2 with Bavachalcone from Aerva lanata at 2.6 A resolution 2zp3 
Carboxylic ester hydrolase, single mutant d49n of bovine pancreatic pla2 enzyme 2zp4 
Carboxylic ester hydrolase, single mutant h48n of bovine pancreatic pla2 enzyme 2zp5 
Carboxylic ester hydrolase, single mutant d49k of bovine pancreatic pla2 enzyme 3bjw 
Crystal Structure of ecarpholin S complexed with suramin 3bp2 
ROLE OF THE N-TERMINUS IN THE INTERACTION OF PANCREATIC PHOSPHOLIPASE A2 WITH AGGREGATED SUBSTRATES. PROPERTIES AND CRYSTAL STRUCTURE OF TRANSAMINATED PHOSPHOLIPASE A2 3cbi 
Crystal structure of the ternary complex of phospholipase A2 with ajmaline and anisic acid at 3.1 A resolution 3cxi 
Structure of BthTX-I complexed with alpha-tocopherol 3cyl 
Crystal structure of Piratoxin I (a myotoxic Lys49-PLA2) complexed with alpha-tocopherol 3dih 
Crystal structure of ammodytin L 3elo 
Crystal Structure of Human Pancreatic Prophospholipase A2 3fg5 
Crystal structure determination of a ternary complex of phospholipase A2 with a pentapetide FLSYK and Ajmaline at 2.5 A resolution 3fo7 
Simultaneous inhibition of anti-coagulation and inflammation: Crystal structure of phospholipase A2 complexed with indomethacin at 1.4 A resolution reveals the presence of the new common ligand binding site 3fvi 
Crystal Structure of Complex of Phospholipase A2 with Octyl Sulfates 3fvj 
Crystal structure of phospholipase A2 1B crystallized in the presence of octyl sulfate 3g8f 
Crystal structure of the complex formed between a group II phospholipase A2 and designed peptide inhibitor carbobenzoxy-dehydro-val-ala-arg-ser at 1.2 A resolution 3g8g 
Crystal structure of phospholipase A2 ammodytoxin A from vipera ammodytes ammodytes 3g8h 
Crystal structure of phospholipase A2 ammodytoxin C from vipera ammodytes ammodytes 3gci 
Crystal Structure of the Complex Formed Between a New Isoform of Phospholipase A2 with C-terminal Amyloid Beta Heptapeptide at 2 A Resolution 3h1x 
Simultaneous inhibition of anti-coagulation and inflammation: Crystal structure of phospholipase A2 complexed with indomethacin at 1.4 A resolution reveals the presence of the new common ligand binding site 3hsw 
Crystal Structure of Porcine Pancreatic Phospholipase A2 in Complex with 2-methoxycyclohexa-2-5-diene-1,4-dione 3hzd 
Crystal structure of bothropstoxin-I (BthTX-I), a PLA2 homologue from Bothrops jararacussu venom 3hzw 
Crystal structure of bothropstoxin-I chemically modified by p-bromophenacyl bromide (BPB) 3i03 
Crystal structure of bothropstoxin-I chemically modified by p-bromophenacyl bromide (BPB) - monomeric form at a high resolution 3i3h 
Crystal structure of Bothropstoxin-I crystallized at 291K 3i3i 
Crystal Structure of Bothropstoxin-I crystallized at 283 K 3iq3 
Crystal Structure of Bothropstoxin-I complexed with polietilene glicol 4000 - crystallized at 283 K 3jq5 
Phospholipase A2 Prevents the Aggregation of Amyloid Beta Peptides: Crystal Structure of the Complex of Phospholipase A2 with Octapeptide Fragment of Amyloid Beta Peptide, Asp-Ala-Glu-Phe-Arg-His-Asp-Ser at 2 A Resolution 3jql 
Crystal Structure of the Complex Formed Between Phospholipase A2 and a Hexapeptide Fragment of Amyloid Beta Peptide, Lys-Leu-Val-Phe-Phe-Ala at 1.2 A Resolution 3jr8 
Crystal Structure of BthTX-II (Asp49-PLA2 from Bothrops jararacussu snake venom) with calcium ions 3jti 
Crystal structure of the complex formed between Phospholipase A2 with beta-amyloid fragment, Lys-Gly-Ala-Ile-Ile-Gly-Leu-Met at 1.8 A resolution 3l30 
Crystal structure of porcine pancreatic phospholipase A2 complexed with dihydroxyberberine 3mlm 
Crystal structure of Bn IV in complex with myristic acid: A Lys49 myotoxic phospholipase A2 from Bothrops neuwiedi venom 3nju 
Crystal structure of the complex of group I phospholipase A2 with 4-Methoxy-benzoicacid at 1.4A resolution 3o4m 
Crystal structure of porcine pancreatic phospholipase A2 in complex with 1,2-dihydroxybenzene 3osh 
Crystal Structure of The Complex of Group 1 Phospholipase A2 With Atropin At 1.5 A Resolution 3p2p 
ENHANCED ACTIVITY AND ALTERED SPECIFICITY OF PHOSPHOLIPASE A2 BY DELETION OF A SURFACE LOOP 3q4y 
Crystal structure of group I phospholipase A2 at 2.3 A resolution in 40% ethanol revealed the critical elements of hydrophobicity of the substrate-binding site 3qlm 
Crystal structure of porcine pancreatic phospholipase A2 in complex with n-hexadecanoic acid 3qnl 
Crystal structure of PrTX-I complexed to Rosmarinic Acid 3r0l 
Crystal structure of crotoxin 3t0r 
Crystal Structure of MjTX-I, a myotoxic Lys49-phospholipase A2 from Bothrops moojeni 3u8b 
Functionally selective inhibition of Group IIA phospholipase A2 reveals a role for vimentin in regulating arachidonic acid metabolism 3u8d 
Functionally selective inhibition of Group IIA phospholipase A2 reveals a role for vimentin in regulating arachidonic acid metabolism 3u8h 
Functionally selective inhibition of Group IIA phospholipase A2 reveals a role for vimentin in regulating arachidonic acid metabolism 3u8i 
Functionally selective inhibition of Group IIA phospholipase A2 reveals a role for vimentin in regulating arachidonic acid metabolism 3ux7 
Crystal structure of a dimeric myotoxic component of the Vipera ammodytes meridionalis venom reveals determinants of myotoxicity and membrane damaging activity 3v9m 
Phospholipase ACII4 from Australian King Brown Snake 3vbz 
Crystal structure of Taipoxin beta subunit isoform 2 3vc0 
Crystal structure of Taipoxin beta subunit isoform 1 4bp2 
CRYSTALLOGRAPHIC REFINEMENT OF BOVINE PRO-PHOSPHOLIPASE A2 AT 1.6 ANGSTROMS RESOLUTION 4dbk 
Crystal structure of porcine pancreatic phospholipase A2 complexed with berberine 4dcf 
Structure of MTX-II from Bothrops brazili 4e4c 
Crystal structure of Notexin at 1.8 A resolution 4eix 
Structural Studies of the ternary complex of Phaspholipase A2 with nimesulide and indomethacin 4fga 
Design of peptide inhibitors of group II phospholipase A2: Crystal structure of the complex of phospholipsae A2 with a designed tripeptide, Ala- Tyr- Lys at 2.3 A resolution 4g5i 
Crystal Structure of Porcine pancreatic PlA2 in complex with DBP 4gfy 
Design of peptide inhibitors of phospholipase A2: crystal Structure of phospholipase A2 complexed with a designed tetrapeptide Val - Ilu- Ala - Lys at 2.7 A resolution 4gld 
Crystal Structure of the complex of type II phospholipase A2 with a designed peptide inhibitor Phe - Leu - Ala - Tyr - Lys at 1.69 A resolution 4h0s 
Crystal structure analysis of a basic phospholipase A2 from Trimeresurus stejnegeri venom 4hg9 
Crystal structure of AhV_bPA, a basic PLA2 from Agkistrodon halys pallas venom 4hmb 
Crystal Structure of the complex of group II phospholipase A2 with a 3-{3-[(Dimethylamino)methyl]-1H-indol-7-yl}propan-1-ol at 2.21 A Resolution 4k06 
Crystal structure of MTX-II from Bothrops brazili venom complexed with polyethylene glycol 4k09 
Crystal structure of BbTX-II from Bothrops brazili venom 4kf3 
Crystal Structure of Myotoxin II (MjTX-II), a myotoxic Lys49-phospholipase A2 from Bothrops moojeni. 4ntw 
Structure of acid-sensing ion channel in complex with snake toxin 4ntx 
Structure of acid-sensing ion channel in complex with snake toxin and amiloride 4nty 
Cesium sites in the crystal structure of acid-sensing ion channel in complex with snake toxin 4o1y 
Crystal structure of Porcine Pancreatic Phospholipase A2 in complex with 1-Naphthaleneacetic acid 4p2p 
AN INDEPENDENT CRYSTALLOGRAPHIC REFINEMENT OF PORCINE PHOSPHOLIPASE A2 AT 2.4 ANGSTROMS RESOLUTION 4qem 
4QEM 4qer 
4QER 4qf7 
4QF7 4qf8 
4QF8 4qgd 
4QGD 4qmc 
4QMC 4rfp 
4RFP 4uy1 
4UY1 4wtb 
4WTB 4yu7 
4YU7 4yv5 
4YV5 4yz7 
4YZ7 5g3m 
5G3M 5g3n 
5G3N 5p2p 
X-RAY STRUCTURE OF PHOSPHOLIPASE A2 COMPLEXED WITH A SUBSTRATE-DERIVED INHIBITOR - Links (links to other resources describing this domain)
-
PROSITE PS00119 INTERPRO IPR016090

