The domain within your query sequence starts at position 11 and ends at position 84; the E-value for the PDZ domain shown below is 1.68e-16.
GPWGFRLQGGKDFNMPLTISRITPGSKAAQSQLSQGDLVVAIDGVNTDTMTHLEAQNKIK SASYNLSLTLQKSK
PDZDomain present in PSD-95, Dlg, and ZO-1/2. |
|---|
| SMART accession number: | SM00228 |
|---|---|
| Description: | Also called DHR (Dlg homologous region) or GLGF (relatively well conserved tetrapeptide in these domains). Some PDZs have been shown to bind C-terminal polypeptides; others appear to bind internal (non-C-terminal) polypeptides. Different PDZs possess different binding specificities. |
| Interpro abstract (IPR001478): | PDZ domains (also known as Discs-large homologous regions (DHR) or GLGF)) are found in diverse signalling proteins in bacteria, yeasts, plants, insects and vertebrates [ (PUBMED:9041651) (PUBMED:9204764) ]. PDZ domains can occur in one or multiple copies and are nearly always found in cytoplasmic proteins. They bind either the carboxyl-terminal sequences of proteins or internal peptide sequences [ (PUBMED:9204764) ]. In most cases, interaction between a PDZ domain and its target is constitutive, with a binding affinity of 1 to 10 microns. However, agonist-dependent activation of cell surface receptors is sometimes required to promote interaction with a PDZ protein. PDZ domain proteins are frequently associated with the plasma membrane, a compartment where high concentrations of phosphatidylinositol 4,5-bisphosphate (PIP2) are found. Direct interaction between PIP2 and a subset of class II PDZ domains (syntenin, CASK, Tiam-1) has been demonstrated. PDZ domains consist of 80 to 90 amino acids comprising six beta-strands (beta-A to beta-F) and two alpha-helices, A and B, compactly arranged in a globular structure. Peptide binding of the ligand takes place in an elongated surface groove as an anti-parallel beta-strand interacts with the beta-B strand and the B helix. The structure of PDZ domains allows binding to a free carboxylate group at the end of a peptide through a carboxylate-binding loop between the beta-A and beta-B strands. |
| GO function: | protein binding (GO:0005515) |
| Family alignment: |
There are 333040 PDZ domains in 224949 proteins in SMART's nrdb database.
Click on the following links for more information.
- Evolution (species in which this domain is found)
-
Taxonomic distribution of proteins containing PDZ domain.
This tree includes only several representative species. The complete taxonomic breakdown of all proteins with PDZ domain is also avaliable.
Click on the protein counts, or double click on taxonomic names to display all proteins containing PDZ domain in the selected taxonomic class.
- Cellular role (predicted cellular role)
-
Cellular role: signalling
Binding / catalysis: protein-binding, C-terminal peptide-bindin - Literature (relevant references for this domain)
-
Primary literature is listed below; Automatically-derived, secondary literature is also avaliable.
- Beebe KD, Shin J, Peng J, Chaudhury C, Khera J, Pei D
- Substrate recognition through a PDZ domain in tail-specific protease.
- Biochemistry. 2000; 39: 3149-55
- Display abstract
Tail-specific protease (Tsp) is a periplasmic enzyme that selectively degrades proteins bearing a nonpolar C-terminus. Its substrate specificity suggests that Tsp may contain a substrate recognition domain, which selectively binds to the nonpolar C-termini of substrate proteins, separate from its catalytic site. In this work, we show that substrate recognition of Tsp is mediated by a PDZ domain, a small protein module that promotes protein-protein interactions by binding to internal or C-terminal sequences of their partner proteins. Partial proteolysis by V8 protease at a single peptide bond immediately N-terminal to the PDZ domain resulted in two distinct and relatively stable fragments and complete loss of catalytic activity. Photoaffinity labeling with a fluorescent nonpolar peptide caused the covalent attachment of the peptide to a single site on the Tsp protein. Systematic deletion mutagenesis of Tsp localized the binding site to amino acids 206-307, a region that completely encompasses the putative PDZ domain (217-301). The isolated PDZ domain (amino acids 206-334) is capable of folding into a well-behaved structure and binds to a nonpolar peptide with a dissociation constant (K(D)) of 1.9 microM, similar to that of the intact Tsp protein. Site-directed mutagenesis of a surface residue at the peptide binding site of the PDZ domain, valine 229, to Glu or Gln resulted in an increase in the K(M) value but had no effect on the k(cat) value. The use of a separate substrate recognition domain such as a PDZ domain may be a general mechanism for achieving selective protein degradation.
- Craven SE, Bredt DS
- PDZ proteins organize synaptic signaling pathways.
- Cell. 1998; 93: 495-8
- Weimbs T, Mostov K, Low SH, Hofmann K
- A model for structural similarity between different SNARE complexes based on sequence relationships.
- Trends Cell Biol. 1998; 8: 260-2
- Cowburn D
- Peptide recognition by PTB and PDZ domains.
- Curr Opin Struct Biol. 1997; 7: 835-8
- Display abstract
Protein tyrosine binding (PTB) and 'post synaptic density disc-large zo-1' (PDZ) domains bind to short peptidic ligands by augmentation of one of the domain's beta sheets and other recognition mechanisms. The two domain classes have a superficial resemblance to each other, even though no sequential homology exists. The structural bases of the interactions are well understood for the few domains now experimentally determined, and ligand-target pairs can probably be identified in favorable cases by analogy with the known domains. For both PTB and PDZ classes, functional activities are still not fully defined: it is possible that these domain classes, along with pleckstrin homology domains, have multiple roles.
- Kim SK
- Polarized signaling: basolateral receptor localization in epithelial cells by PDZ-containing proteins.
- Curr Opin Cell Biol. 1997; 9: 853-9
- Display abstract
Extracellular signals are normally presented to one surface of epithelial cells and to one end of neurons, and so neuronal and epithelial cell signaling is inherently polarized. Another aspect of signaling polarity is that receptors are often asymmetrically distributed on the surfaces of polarized cells. Recent evidence from studies of Caenorhabditis elegans shows that signaling polarity plays an important role in development. The underlying mesoderm induces the overlying ectoderm to form the vulva, and asymmetric distribution of the signal receptor on the basolateral surface of the epithelium is crucial for this signaling. In neurons, the localization of neurotransmitter receptors and ion channels at synapses allows neurons to be exquisitely sensitive to synaptic inputs. Exciting recent reports suggest that receptor localization to neuronal synapses and the basolateral membrane domains of epithelia may involve a common molecular mechanism involving localization by PDZ-containing proteins.
- Kornau HC, Seeburg PH, Kennedy MB
- Interaction of ion channels and receptors with PDZ domain proteins.
- Curr Opin Neurobiol. 1997; 7: 368-73
- Display abstract
The complex anatomy of neurons demands a high degree of functional organization. Therefore, membrane receptors and ion channels are often localized to selected subcellular sites and coupled to specific signal transduction machineries. PDZ domains have come into focus as protein interaction modules that mediate the binding of a class of submembraneous proteins to membrane receptors and ion channels and thus subserve these organizational aspects. The structures of two PDZ domains have been resolved, which has led to a structural understanding of the specificity of interactions of various PDZ domains with their respective partners. The functional implications of PDZ domain interactions are now being addressed in vitro and in vivo.
- Ponting CP
- Evidence for PDZ domains in bacteria, yeast, and plants.
- Protein Sci. 1997; 6: 464-8
- Display abstract
Several dozen signaling proteins are now known to contain 80-100 residue repeats, called PDZ (or DHR or GLGF) domains, several of which interact with the C-terminal tetrapeptide motifs X-Ser/Thr-X-Val-COO- of ion channels and/or receptors. PDZ domains have previously been noted only in mammals, flies, and worms, suggesting that the primordial PDZ domain arose relatively late in eukaryotic evolution. Here, techniques of sequence analysis-including local alignment, profile, and motif database searches-indicate that PDZ domain homologues are present in yeast, plants, and bacteria. It is suggested that two PDZ domains occur in bacterial high-temperature requirement A (htrA) and one in tail-specific protease (tsp) homologues, and that a yeast htrA homologue contains four PDZ domains. Sequence comparisons suggest that the spread of PDZ domains in these diverse organisms may have occurred via horizontal gene transfer. The known affinity of Escherichia coli tsp for C-terminal polypeptides is proposed to be mediated by its PDZ-like domain, in a similar manner to the binding of C-terminal polypeptides by animal PDZ domains.
- Ponting CP, Phillips C, Davies KE, Blake DJ
- PDZ domains: targeting signalling molecules to sub-membranous sites.
- Bioessays. 1997; 19: 469-79
- Display abstract
PDZ (also called DHR or GLGF) domains are found in diverse membrane-associated proteins including members of the MAGUK family of guanylate kinase homologues, several protein phosphatases and kinases, neuronal nitric oxide synthase, and several dystrophin-associated proteins, collectively known as syntrophins. Many PDZ domain-containing proteins appear to be localised to highly specialised submembranous sites, suggesting their participation in cellular junction formation, receptor or channel clustering, and intracellular signalling events. PDZ domains of several MAGUKs interact with the C-terminal polypeptides of a subset of NMDA receptor subunits and/or with Shaker-type K+ channels. Other PDZ domains have been shown to bind similar ligands of other transmembrane receptors. Recently, the crystal structures of PDZ domains, with and without ligand, have been determined. These demonstrate the mode of ligand-binding and the structural bases for sequence conservation among diverse PDZ domains.
- Ranganathan R, Ross EM
- PDZ domain proteins: scaffolds for signaling complexes.
- Curr Biol. 1997; 7: 7703-7703
- Display abstract
InaD, a Drosophila photoreceptor scaffolding protein, assembles multiple signal-transducing proteins at the membrane via its five PDZ domains, enhancing speed and efficiency of vision. Extensive conservation of PDZ domains suggests that these motifs have a general role in organizing diverse signaling complexes.
- Songyang Z et al.
- Recognition of unique carboxyl-terminal motifs by distinct PDZ domains.
- Science. 1997; 275: 73-7
- Display abstract
The oriented peptide library technique was used to investigate the peptide-binding specificities of nine PDZ domains. Each PDZ domain selected peptides with hydrophobic residues at the carboxyl terminus. Individual PDZ domains selected unique optimal motifs defined primarily by the carboxyl terminal three to seven residues of the peptides. One family of PDZ domains, including those of the Discs Large protein, selected peptides with the consensus motif Glu-(Ser/Thr)-Xxx-(Val/Ile) (where Xxx represents any amino acid) at the carboxyl terminus. In contrast, another family of PDZ domains, including those of LIN-2, p55, and Tiam-1, selected peptides with hydrophobic or aromatic side chains at the carboxyl terminal three residues. On the basis of crystal structures of the PSD-95-3 PDZ domain, the specificities observed with the peptide library can be rationalized.
- Doyle DA, Lee A, Lewis J, Kim E, Sheng M, MacKinnon R
- Crystal structures of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ.
- Cell. 1996; 85: 1067-76
- Display abstract
Modular PDZ domains, found in many cell junction-associated proteins, mediate the clustering of membrane ion channels by binding to their C-terminus. The X-ray crystallographic structures of the third PDZ domain from the synaptic protein PSD-95 in complex with and in the absence of its peptide ligand have been determined at 1.8 angstroms and 2.3 angstroms resolution, respectively. The structures reveal that a four-residue C-terminal stretch (X-Thr/Ser-X-Val-COO(-)) engages the PDZ domain through antiparallel main chain interactions with a beta sheet of the domain. Recognition of the terminal carboxylate group of the peptide is conferred by a cradle of main chain amides provided by a Gly-Leu-Gly-Phe loop as well as by an arginine side chain. Specific side chain interactions and a prominent hydrophobic pocket explain the selective recognition of the C-terminal consensus sequence.
- Fanning AS, Anderson JM
- Protein-protein interactions: PDZ domain networks.
- Curr Biol. 1996; 6: 1385-8
- Display abstract
PDZ domains can dimerize or bind to the carboxyl termini of unrelated proteins. Crystallographic studies demonstrate the structural basis for these interactions, which contribute to the ability of PDZ domains to create networks associated with the plasma membrane.
- Gomperts SN
- Clustering membrane proteins: It's all coming together with the PSD-95/SAP90 protein family.
- Cell. 1996; 84: 659-62
- MoraisCabral JH et al.
- Crystal structure of a PDZ domain.
- Nature. 1996; 382: 649-52
- Display abstract
PDZ domains (also known as DHR domains or GLGF repeats) are approximately 90-residue repeats found in a number of proteins implicated in ion-channel and receptor clustering, and the linking of receptors to effector enzymes. PDZ domains are protein-recognition modules; some recognize proteins containing the consensus carboxy-terminal tripeptide motif S/TXV with high specificity. Other PDZ domains form homotypic dimers: the PDZ domain of the neuronal enzyme nitric oxide synthase binds to the PDZ domain of PSD-95, an interaction that has been implicated in its synaptic association. Here we report the crystal structure of the third PDZ domain of the human homologue of the Drosophila discs-large tumour-suppressor gene product, DlgA. It consists of a five-stranded antiparallel beta-barrel flanked by three alpha-helices. A groove runs over the surface of the domain, ending in a conserved hydrophobic pocket and a buried arginine; we suggest that this is the binding site for the C-terminal peptide.
- Kennedy MB
- Origin of PDZ (DHR, GLGF) domains.
- Trends Biochem Sci. 1995; 20: 350-350
- Ponting CP, Phillips C
- DHR domains in syntrophins, neuronal NO synthases and other intracellular proteins.
- Trends Biochem Sci. 1995; 20: 102-3
- Metabolism (metabolic pathways involving proteins which contain this domain)
-
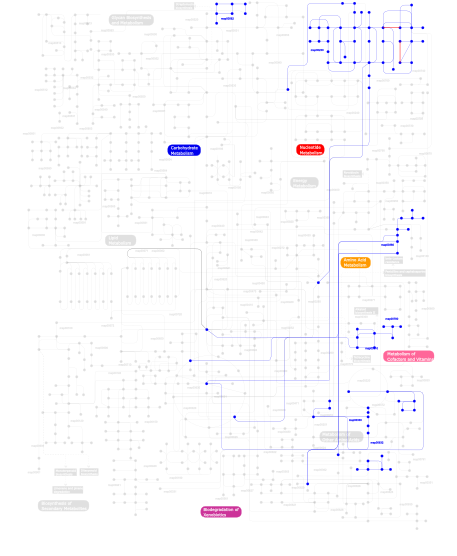
Click the image to view the interactive version of the map in iPath% proteins involved KEGG pathway ID Description 20.11 map04530 Tight junction 11.11 map02020 Two-component system - General 6.13 map04520 Adherens junction 5.56 map04810 Regulation of actin cytoskeleton 5.56 map04360 Axon guidance 4.60 map04310 Wnt signaling pathway 4.60 map04916 Melanogenesis 4.60 map05217 Basal cell carcinoma 4.60 map05210 Colorectal cancer 4.60 map04330 Notch signaling pathway 2.87 map03050 Proteasome 2.30 map03090 Type II secretion system 2.11 map04670 Leukocyte transendothelial migration 1.92 map05050 Dentatorubropallidoluysian atrophy (DRPLA) 1.92 map04080 Neuroactive ligand-receptor interaction 1.53  map00230
map00230Purine metabolism 1.34 map05120 Epithelial cell signaling in Helicobacter pylori infection 1.34 map04662 B cell receptor signaling pathway 1.34 map04660 T cell receptor signaling pathway 1.34 map04540 Gap junction 1.15 map05222 Small cell lung cancer 1.15 map04020 Calcium signaling pathway 1.15 map04010 MAPK signaling pathway 1.15  map00380
map00380Tryptophan metabolism 1.15  map00330
map00330Arginine and proline metabolism 1.15 map04730 Long-term depression 0.57  map00780
map00780Biotin metabolism 0.57  map00310
map00310Lysine degradation 0.38  map00632
map00632Benzoate degradation via CoA ligation 0.38  map00562
map00562Inositol phosphate metabolism 0.19 map04920 Adipocytokine signaling pathway 0.19 map05214 Glioma 0.19 map03060 Protein export 0.19 map04910 Insulin signaling pathway 0.19 map05215 Prostate cancer 0.19 map04012 ErbB signaling pathway 0.19 map04150 mTOR signaling pathway 0.19 map05221 Acute myeloid leukemia 0.19 map04930 Type II diabetes mellitus This information is based on mapping of SMART genomic protein database to KEGG orthologous groups. Percentage points are related to the number of proteins with PDZ domain which could be assigned to a KEGG orthologous group, and not all proteins containing PDZ domain. Please note that proteins can be included in multiple pathways, ie. the numbers above will not always add up to 100%.
- Structure (3D structures containing this domain)
3D Structures of PDZ domains in PDB
PDB code Main view Title 1b8q 
SOLUTION STRUCTURE OF THE EXTENDED NEURONAL NITRIC OXIDE SYNTHASE PDZ DOMAIN COMPLEXED WITH AN ASSOCIATED PEPTIDE 1be9 
THE THIRD PDZ DOMAIN FROM THE SYNAPTIC PROTEIN PSD-95 IN COMPLEX WITH A C-TERMINAL PEPTIDE DERIVED FROM CRIPT. 1bfe 
THE THIRD PDZ DOMAIN FROM THE SYNAPTIC PROTEIN PSD-95 1d5g 
SOLUTION STRUCTURE OF THE PDZ2 DOMAIN FROM HUMAN PHOSPHATASE HPTP1E COMPLEXED WITH A PEPTIDE 1fc6 
PHOTOSYSTEM II D1 C-TERMINAL PROCESSING PROTEASE 1fc7 
PHOTOSYSTEM II D1 C-TERMINAL PROCESSING PROTEASE 1fc9 
PHOTOSYSTEM II D1 C-TERMINAL PROCESSING PROTEASE 1fcf 
PHOTOSYSTEM II D1 C-TERMINAL PROCESSING PROTEASE 1g9o 
FIRST PDZ DOMAIN OF THE HUMAN NA+/H+ EXCHANGER REGULATORY FACTOR 1gm1 
Second PDZ Domain (PDZ2) of PTP-BL 1gq4 
STRUCTURAL DETERMINANTS OF THE NHERF INTERACTION WITH BETA2AR AND PDGFR 1gq5 
Structural Determinants of the NHERF Interaction with beta2-AR and PDGFR 1i16 
STRUCTURE OF INTERLEUKIN 16: IMPLICATIONS FOR FUNCTION, NMR, 20 STRUCTURES 1i92 
STRUCTURAL BASIS OF THE NHERF PDZ1-CFTR INTERACTION 1ihj 
Crystal Structure of the N-terminal PDZ domain of InaD in complex with a NorpA C-terminal peptide 1iu0 
The first PDZ domain of PSD-95 1iu2 
The first PDZ domain of PSD-95 1kef 
PDZ1 of SAP90 1kwa 
HUMAN CASK/LIN-2 PDZ DOMAIN 1ky9 
Crystal Structure of DegP (HtrA) 1l6o 
XENOPUS DISHEVELLED PDZ DOMAIN 1lcy 
Crystal Structure of the Mitochondrial Serine Protease HtrA2 1m5z 
The PDZ7 of Glutamate Receptor Interacting Protein Binds to its Target via a Novel Hydrophobic Surface Area 1mc7 
Solution Structure of mDvl1 PDZ domain 1mfg 
The Structure of ERBIN PDZ domain bound to the Carboxy-terminal tail of the ErbB2 Receptor 1mfl 
The Structure of ERBIN PDZ domain bound to the Carboxy-terminal tail of the ErbB2 Receptor 1n7e 
Crystal structure of the sixth PDZ domain of GRIP1 1n7f 
Crystal structure of the sixth PDZ domain of GRIP1 in complex with liprin C-terminal peptide 1n7t 
ERBIN PDZ domain bound to a phage-derived peptide 1n99 
CRYSTAL STRUCTURE OF THE PDZ TANDEM OF HUMAN SYNTENIN 1nf3 
Structure of Cdc42 in a complex with the GTPase-binding domain of the cell polarity protein, Par6 1nte 
CRYSTAL STRUCTURE ANALYSIS OF THE SECOND PDZ DOMAIN OF SYNTENIN 1obx 
Crystal structure of the complex of PDZ2 of syntenin with an interleukin 5 receptor alpha peptide. 1oby 
Crystal structure of the complex of PDZ2 of syntenin with a syndecan-4 peptide. 1obz 
Crystal structure of the complex of the PDZ tandem of syntenin with an interleukin 5 receptor alpha peptide. 1ozi 
The alternatively spliced PDZ2 domain of PTP-BL 1p1d 
Structural Insights into the Inter-domain Chaperoning of Tandem PDZ Domains in Glutamate Receptor Interacting Proteins 1p1e 
Structural Insights into the Inter-domain Chaperoning of Tandem PDZ Domains in Glutamate Receptor Interacting Proteins 1pdr 
CRYSTAL STRUCTURE OF THE THIRD PDZ DOMAIN FROM THE HUMAN HOMOLOG OF DISCS LARGE PROTEIN 1q3o 
Crystal structure of the Shank PDZ-ligand complex reveals a class I PDZ interaction and a novel PDZ-PDZ dimerization 1q3p 
Crystal structure of the Shank PDZ-ligand complex reveals a class I PDZ interaction and a novel PDZ-PDZ dimerization 1q7x 
Solution structure of the alternatively spliced PDZ2 domain (PDZ2b) of PTP-Bas (hPTP1E) 1qau 
UNEXPECTED MODES OF PDZ DOMAIN SCAFFOLDING REVEALED BY STRUCTURE OF NNOS-SYNTROPHIN COMPLEX 1qav 
Unexpected Modes of PDZ Domain Scaffolding Revealed by Structure of NNOS-Syntrophin Complex 1qlc 
Solution structure of the second PDZ domain of Postsynaptic Density-95 1r6j 
Ultrahigh resolution Crystal Structure of syntenin PDZ2 1rgr 
Cyclic Peptides Targeting PDZ Domains of PSD-95: Structural Basis for Enhanced Affinity and Enzymatic Stability 1rgw 
Solution Structure of ZASP's PDZ domain 1ry4 
NMR Structure of the CRIB-PDZ module of Par-6 1rzx 
Crystal Structure of a Par-6 PDZ-peptide Complex 1sot 
Crystal Structure of the DegS stress sensor 1soz 
Crystal Structure of DegS protease in complex with an activating peptide 1t2m 
Solution Structure Of The Pdz Domain Of AF-6 1te0 
Structural analysis of DegS, a stress sensor of the bacterial periplasm 1tp3 
PDZ3 domain of PSD-95 protein complexed with KKETPV peptide ligand 1tp5 
Crystal structure of PDZ3 domain of PSD-95 protein complexed with a peptide ligand KKETWV 1tq3 
Higher resolution crystal structure of the third PDZ domain of post synaptic PSD-95 protein 1u37 
Auto-inhibition Mechanism of X11s/Mints Family Scaffold Proteins Revealed by the Closed Conformation of the Tandem PDZ Domains 1u38 
Auto-inhibition Mechanism of X11s/Mints Family Scaffold Proteins Revealed by the Closed Conformation of the Tandem PDZ Domains 1u39 
Auto-inhibition Mechanism of X11s/Mints Family Scaffold Proteins Revealed by the Closed Conformation of the Tandem PDZ Domains 1u3b 
Auto-inhibition Mechanism of X11s/Mints Family Scaffold Proteins Revealed by the Closed Conformation of the Tandem PDZ Domains 1uep 
Solution Structure of The Third PDZ Domain of Human Atrophin-1 Interacting Protein 1 (KIAA0705 Protein) 1ueq 
Solution Structure of The First PDZ domain of Human Atrophin-1 Interacting Protein 1 (KIAA0705 protein) 1uew 
Solution Structure of The forth PDZ Domain of Human Atrophin-1 Interacting Protein 1 (KIAA0705 Protein) 1uez 
Solution structure of the first PDZ domain of human KIAA1526 protein 1uf1 
Solution structure of the second PDZ domain of human KIAA1526 protein 1ufx 
Solution structure of the third PDZ domain of human KIAA1526 protein 1uhp 
Solution structure of RSGI RUH-005, a PDZ domain in human cDNA, KIAA1095 1uit 
Solution structure of RSGI RUH-006, The third PDZ domain OF hDlg5 (KIAA0583) protein [Homo sapiens] 1ujd 
Solution Structure of RSGI RUH-003, a PDZ domain of hypothetical KIAA0559 protein from human cDNA 1uju 
Solution structure of the fourth PDZ domain of human scribble (KIAA0147 protein) 1ujv 
Solution structure of the second PDZ domain of human membrane associated guanylate kinase inverted-2 (MAGI-2) 1um1 
Solution Structure of RSGI RUH-007, PDZ domain in Human cDNA 1um7 
Solution structure of the third PDZ domain of synapse-associated protein 102 1v1t 
Crystal structure of the PDZ tandem of human syntenin with TNEYKV peptide 1v5l 
Solution structure of PDZ domain of mouse Alpha-actinin-2 associated LIM protein 1v5q 
Solution Structure of the PDZ Domain from Mouse Glutamate Receptor Interacting Protein 1A-L (GRIP1) Homolog 1v62 
Solution structure of the 3rd PDZ domain of GRIP2 1v6b 
Solution structure of the third PDZ domain of mouse harmonin 1va8 
Solution structure of the PDZ domain of Pals1 protein 1vae 
Solution structure of the PDZ domain of mouse Rhophilin-2 1vb7 
Solution structure of the PDZ domain of PDZ and LIM domain 2 1vcw 
Crystal structure of DegS after backsoaking the activating peptide 1vj6 
PDZ2 from PTP-BL in complex with the C-terminal ligand from the APC protein 1w9e 
Crystal structure of the PDZ tandem of human syntenin in complex with TNEFYF peptide 1w9o 
Crystal structure of the PDZ tandem of human syntenin in complex with TNEYYV peptide 1w9q 
Crystal structure of the PDZ tandem of human syntenin in complex with TNEFAF peptide 1wf7 
Solution structure of the PDZ domain of Enigma homologue protein 1wf8 
Solution structure of the PDZ domain of Spinophilin/NeurabinII protein 1wfg 
PDZ domain of human RIM2B 1wfv 
Solution structure of the fifth PDZ domain of human membrane associated guanylate kinase inverted-2 (KIAA0705 protein) 1wg6 
Solution structure of PDZ domain in protein XP_110852 1wh1 
Solution structure of the fourth PDZ domain of KIAA1095 protein 1wha 
Solution structure of the second PDZ domain of human scribble (KIAA0147 protein). 1whd 
Solution structure of the PDZ domain of RGS3 1wi2 
Solution structure of the PDZ domain from RIKEN cDNA 2700099C19 1wi4 
Solution structure of the PDZ domain of syntaxin binding protein 4 1wif 
The solution structure of RSGI RUH-020, a PDZ domain of hypothetical protein from mouse 1wjl 
Solution structure of PDZ domain of mouse Cypher protein 1x45 
Solution structure of the first PDZ domain of amyloid beta A4 precursor protein-binding family A, member 1 1x5n 
Solution structure of the second PDZ domain of harmonin protein 1x5q 
Solution structure of the first PDZ domain of scribble homolog protein (hScrib) 1x5r 
Solution structure of the fourth PDZ domain of Glutamate receptor interacting protein 2 1x6d 
Solution structures of the PDZ domain of human Interleukin-16 1x8s 
Structure of the Par-6 PDZ domain with a Pals1 internal ligand 1xz9 
Structure of AF-6 PDZ domain 1y7n 
Solution structure of the second PDZ domain of the human neuronal adaptor X11alpha 1y8t 
Crystal Structure of RV0983 from Mycobacterium tuberculosis- Proteolytically active form 1ybo 
Crystal structure of the PDZ tandem of human syntenin with syndecan peptide 1z86 
Solution structure of the PDZ domain of alpha-syntrophin 1z87 
solution structure of the split PH-PDZ Supramodule of alpha-Syntrophin 1zok 
PDZ1 Domain Of Synapse Associated Protein 97 1zub 
Solution Structure of the RIM1alpha PDZ Domain in Complex with an ELKS1b C-terminal Peptide 2ain 
Solution structure of the AF-6 PDZ domain complexed with the C-terminal peptide from the Bcr protein 2awu 
Synapse associated protein 97 PDZ2 domain variant C378G 2aww 
Synapse associated protein 97 PDZ2 domain variant C378G with C-terminal GluR-A peptide 2awx 
Synapse associated protein 97 PDZ2 domain variant C378S 2byg 
2nd PDZ Domain of Discs Large Homologue 2 2cs5 
Solution structure of PDZ domain of Protein tyrosine phosphatase, non-receptor type 4 2csj 
Solution structure of N-terminal PDZ domain from mouse TJP2 2css 
Solution structure of the PDZ domain of human KIAA0340 protein 2d8i 
Solution structure of the PDZ domain of T-cell lymphoma invasion and metastasis 1 varian 2d90 
Solution structure of the third PDZ domain of PDZ domain containing protein 1 2d92 
Solution structure of the fifth PDZ domain of InaD-like protein 2daz 
Solution structure of the 7th PDZ domain of InaD-like protein 2db5 
Solution structure of the first PDZ domain of InaD-like protein 2dc2 
Solution Structure of PDZ Domain 2djt 
Solution structures of the PDZ domain of human unnamed protein product 2dkr 
Solution structure of the PDZ domain from human Lin-7 homolog B 2dls 
Solution structure of the PDZ domain of human Rho guanine nucleotide exchange factor 11 2dlu 
Solution structure of the second PDZ domain of human InaD-like protein 2dm8 
Solution structure of the eighth PDZ domain of human InaD-like protein 2dmz 
Solution structure of the third PDZ domain of human InaD-like protein 2e7k 
Solution structure of the PDZ domain from Human MAGUK p55 subfamily member 2 2eaq 
Crystal structure of PDZ domain of KIAA0858 (LIM), MS0793 from Homo sapiens 2edp 
Solution structure of the PDZ domain from human Shroom family member 4 2edv 
Solution structure of the PDZ domain from human FERM and PDZ domain containing 1 2edz 
Solution structures of the PDZ domain of mus musculus PDZ domain-containing protein 1 2eeg 
Solution Structure of PDZ domain of PDZ and LIM domain protein 2eeh 
Solution Structure of First PDZ domain of PDZ Domain Containing Protein 7 2eei 
Solution Structure of Second PDZ domain of PDZ Domain Containing Protein 1 2eej 
Solution Structure of Fourth PDZ domain of PDZ Domain Containing Protein 1 2egk 
Crystal Structure of Tamalin PDZ-Intrinsic Ligand Fusion Protein 2egn 
Crystal Structure of Tamalin PDZ Domain in Complex with mGluR5 C-terminal Peptide 2ego 
Crystal Structure of Tamalin PDZ Domain 2ehr 
Solution structure of the sixth PDZ domain of human InaD-like protein 2ejy 
Solution structure of the p55 PDZ T85C domain complexed with the glycophorin C F127C peptide 2eno 
Solution structure of the PDZ domain from human Synaptojanin 2 binding protein 2ev8 
Solution structure of the erythroid p55 PDZ domain 2exg 
Making Protein-Protein Interactions Drugable: Discovery of Low-Molecular-Weight Ligands for the AF6 PDZ Domain 2f0a 
Crystal Structure of Monomeric Uncomplexed form of Xenopus dishevelled PDZ domain 2f5y 
Crystal Structure of the PDZ Domain from Human RGS-3 2fcf 
The crystal structure of the 7th PDZ domain of MPDZ (MUPP-1) 2fe5 
The Crystal Structure of the Second PDZ Domain of Human DLG3 2fn5 
NMR Structure of the Neurabin PDZ domain (502-594) 2fne 
The crystal structure of the 13th PDZ domain of MPDZ 2g2l 
Crystal Structure of the Second PDZ Domain of SAP97 in Complex with a GluR-A C-terminal Peptide 2g5m 
Spinophilin PDZ domain 2gzv 
The cystal structure of the PDZ domain of human PICK1 (CASP TARGET) 2h2b 
Crystal Structure of ZO-1 PDZ1 Bound to a Phage-Derived Ligand (WRRTTYL) 2h2c 
Crystal Structure of ZO-1 PDZ1 Bound to a Phage-Derived Ligand (WRRTTWV) 2h3l 
Crystal Structure of ERBIN PDZ 2h3m 
Crystal Structure of ZO-1 PDZ1 2he2 
Crystal structure of the 3rd PDZ domain of human discs large homologue 2, DLG2 2he4 
The crystal structure of the second PDZ domain of human NHERF-2 (SLC9A3R2) interacting with a mode 1 PDZ binding motif 2hga 
Solution NMR Structure of Conserved protein MTH1368, Northeast Structural Genomics Consortium Target TT821A 2i04 
X-ray crystal structure of MAGI-1 PDZ1 bound to the C-terminal peptide of HPV18 E6 2i0i 
X-ray crystal structure of Sap97 PDZ3 bound to the C-terminal peptide of HPV18 E6 2i0l 
X-ray crystal structure of Sap97 PDZ2 bound to the C-terminal peptide of HPV18 E6. 2i1n 
Crystal structure of the 1st PDZ domain of Human DLG3 2iwn 
3rd PDZ domain of Multiple PDZ Domain Protein MPDZ (CASP Target) 2iwo 
12th PDZ domain of Multiple PDZ Domain Protein MPDZ (CASP Target) 2iwp 
12th PDZ domain of Multiple PDZ Domain Protein MPDZ (CASP Target) 2iwq 
7th PDZ domain of Multiple PDZ Domain Protein MPDZ 2jik 
Crystal structure of PDZ domain of Synaptojanin-2 binding protein 2jil 
Crystal structure of 2nd PDZ domain of glutamate receptor interacting protein-1 (GRIP1) 2jin 
Crystal structure of PDZ domain of Synaptojanin-2 binding protein 2joa 
HtrA1 bound to an optimized peptide: NMR assignment of PDZ domain and ligand resonances 2jre 
C60-1, a PDZ domain designed using statistical coupling analysis 2jwe 
Solution structure of the second PDZ domain from human zonula occludens-1: A dimeric form with 3D domain swapping 2jxo 
Structure of the second PDZ domain of NHERF-1 2k1z 
Solution structure of Par-3 PDZ3 2k20 
Solution structure of Par-3 PDZ3 in complex with PTEN peptide 2ka9 
Solution structure of PSD-95 PDZ12 complexed with cypin peptide 2kaw 
NMR structure of the mDvl1 PDZ domain in complex with its inhibitor 2kbs 
Solution structure of harmonin PDZ2 in complex with the carboxyl tail peptide of cadherin23 2kg2 
Solution structure of a PDZ protein 2kjd 
Solution structure of extended PDZ2 domain from NHERF1 (150-270) 2kjk 
Solution structure of the second domain of the listeria protein Lin2157, Northeast Structural Genomics Consortium target Lkr136b 2kjp 
Solution structure of protein YlbL (BSU15050) from Bacillus subtilis, Northeast Structural Genomics Consortium target sr713a 2kl1 
Solution structure of GtR34C from Geobacillus thermodenitrificans. Northeast Structural Genomics Consortium Target GtR34C 2koh 
NMR structure of mouse Par3-PDZ3 in complex with VE-Cadherin C-terminus 2koj 
Solution structure of mouse Par-3 PDZ2 (residues 450-558) 2kom 
Solution structure of humar Par-3b PDZ2 (residues 451-549) 2kpk 
MAGI-1 PDZ1 2kpl 
MAGI-1 PDZ1 / E6CT 2kqf 
Solution structure of MAST205-PDZ complexed with the C-terminus of a rabies virus G protein 2krg 
Solution Structure of human sodium/ hydrogen exchange regulatory factor 1(150-358) 2kv8 
Solution structure ofRGS12 PDZ domain 2kyl 
Solution structure of MAST2-PDZ complexed with the C-terminus of PTEN 2l4s 
Promiscuous Binding at the Crossroads of Numerous Cancer Pathways: Insight from the Binding of GIP with Glutaminase L 2l4t 
GIP/Glutaminase L peptide complex 2l97 
Solution structure of HtrA PDZ domain from Streptococcus pneumoniae 2la8 
Solution structure of INAD PDZ5 complexed with Kon-tiki peptide 2lc6 
Solution structure of Par-6 Q144C/L164C 2lc7 
Solution structure of the isolated Par-6 PDZ domain 2lob 
PDZ Domain of CAL (Cystic Fibrosis Transmembrane Regulator-Associated Ligand) 2lui 
Structure of the PICK PDZ domain in complex with the DAT C-terminal 2m0t 
Structural characterization of the extended PDZ1 domain from NHERF1 2m0u 
Complex structure of C-terminal CFTR peptide and extended PDZ1 domain from NHERF1 2m0v 
Complex structure of C-terminal CFTR peptide and extended PDZ2 domain from NHERF1 2m0z 
cis form of a photoswitchable PDZ domain crosslinked with an azobenzene derivative 2m10 
trans form of a photoswitchable PDZ domain crosslinked with an azobenzene derivative 2m3m 
Solution structure of a complex consisting of hDlg/SAP-97 residues 318-406 and HPV51 oncoprotein E6 residues 141-151 2mho 
Solution State Structure PSD-95 PDZ1 with 5HT2C Receptor peptide 2mx6 
2MX6 2n7p 
2N7P 2nb4 
2NB4 2o2t 
The crystal structure of the 1st PDZ domain of MPDZ 2ocs 
The crystal structure of the first PDZ domain of human NHERF-2 (SLC9A3R2) 2ogp 
Solution structure of the second PDZ domain of Par-3 2omj 
solution structure of LARG PDZ domain 2opg 
The crystal structure of the 10th PDZ domain of MPDZ 2oqs 
Structure of the hDLG/SAP97 PDZ2 in complex with HPV-18 papillomavirus E6 peptide 2os6 
Solution structure of LARG PDZ domain in complex with C-terminal octa-peptide of Plexin B1 2osg 
Solution Structure and Binding Property of the Domain-swapped Dimer of ZO2PDZ2 2ozf 
The crystal structure of the 2nd PDZ domain of the human NHERF-1 (SLC9A3R1) 2p3w 
Crystal Structure of the HtrA3 PDZ Domain Bound to a Phage-Derived Ligand (FGRWV) 2pa1 
Structure of the PDZ domain of human PDLIM2 bound to a C-terminal extension from human beta-tropomyosin 2pdz 
SOLUTION STRUCTURE OF THE SYNTROPHIN PDZ DOMAIN IN COMPLEX WITH THE PEPTIDE GVKESLV, NMR, 15 STRUCTURES 2pkt 
Crystal structure of the human CLP-36 (PDLIM1) bound to the C-terminal peptide of human alpha-actinin-1 2pku 
Solution structure of PICK1 PDZ in complex with the carboxyl tail peptide of GluR2 2pnt 
Crystal structure of the PDZ domain of human GRASP (GRP1) in complex with the C-terminal peptide of the metabotropic glutamate receptor type 1 2pzd 
Crystal Structure of the HtrA2/Omi PDZ Domain Bound to a Phage-Derived Ligand (WTMFWV) 2q3g 
Structure of the PDZ domain of human PDLIM7 bound to a C-terminal extension from human beta-tropomyosin 2q9v 
Crystal structure of the C890S mutant of the 4th PDZ domain of human membrane associated guanylate kinase 2qbw 
The crystal structure of PDZ-Fibronectin fusion protein 2qg1 
Crystal structure of the 11th PDZ domain of MPDZ (MUPP1) 2qkt 
Crystal Structure of the 5th PDZ domain of InaD 2qku 
The 5th PDZ Domain of InaD in 10mM DTT 2qkv 
Crystal Structure of the C645S Mutant of the 5th PDZ Domain of InaD 2qt5 
Crystal Structure of GRIP1 PDZ12 in Complex with the Fras1 Peptide 2r3y 
Crystal structure of the DegS protease in complex with the YWF activating peptide 2r4h 
Crystal structure of a C1190S mutant of the 6th PDZ domain of human membrane associated guanylate kinase 2rcz 
Structure of the second PDZ domain of ZO-1 2rey 
Crystal structure of the PDZ domain of human dishevelled 2 (homologous to Drosophila dsh) 2rrm 
Interplay between phosphatidyl-inositol-phosphates and claudins upon binding to the 1st PDZ domain of zonula occludens 1 2uzc 
Structure of human PDLIM5 in complex with the C-terminal peptide of human alpha-actinin-1 2v1w 
Crystal structure of human LIM protein RIL (PDLIM4) PDZ domain bound to the C-terminal peptide of human alpha-actinin-1 2v90 
Crystal structure of the 3rd PDZ domain of intestine- and kidney- enriched PDZ domain IKEPP (PDZD3) 2vph 
Crystal structure of the human protein tyrosine phosphatase, non- receptor type 4, PDZ domain 2vrf 
CRYSTAL STRUCTURE OF THE HUMAN BETA-2-SYNTROPHIN PDZ DOMAIN 2vsp 
Crystal structure of the fourth PDZ domain of PDZ domain-containing protein 1 2vsv 
Crystal structure of the PDZ domain of human rhophilin-2 2vwr 
Crystal structure of the second pdz domain of numb-binding protein 2 2vz5 
Structure of the PDZ domain of Tax1 (human T-cell leukemia virus type I) binding protein 3 2w4f 
CRYSTAL STRUCTURE OF THE FIRST PDZ DOMAIN OF HUMAN SCRIB1 2w7r 
Structure of the PDZ domain of Human Microtubule associated serine- threonine kinase 4 2wl7 
Crystal structure of the PSD93 PDZ1 domain 2x7z 
Crystal Structure of the SAP97 PDZ2 I342W C378A mutant protein domain 2xkx 
Single particle analysis of PSD-95 in negative stain 2yt7 
Solution structure of the PDZ domain of Amyloid beta A4 precursor protein-binding family A member 3 2yt8 
Solution structure of the PDZ domain of Amyloid beta A4 precursor protein-binding family A member 3 (Neuron- specific X11L2 protein) (Neuronal Munc18-1-interacting protein 3) (Mint-3) (Adapter protein X11gamma) 2ytw 
Solution structure of the PDZ-domain of human protease HTRA 1 precursor 2yub 
Solution structure of the PDZ domain from mouse LIM domain kinase 2yuy 
Solution Structure of PDZ domain of Rho GTPase Activating Protein 21 2z17 
Crystal sturcture of PDZ domain from human Pleckstrin homology, Sec7 2z9i 
Crystal structure of RV0983 from Mycobacterium tuberculosis- Proteolytically active form 2zle 
Cryo-EM structure of DegP12/OMP 2zpm 
Crystal structure analysis of PDZ domain B 3axa 
Crystal structure of afadin PDZ domain in complex with the C-terminal peptide from nectin-3 3b76 
Crystal structure of the third PDZ domain of human ligand-of-numb protein-X (LNX1) in complex with the C-terminal peptide from the coxsackievirus and adenovirus receptor 3bpu 
Crystal structure of the 3rd PDZ domain of human membrane associated guanylate kinase, C677S and C709S double mutant 3cbx 
The Dvl2 PDZ Domain in Complex with the C1 Inhibitory Peptide 3cby 
The Dvl2 PDZ Domain in Complex with the N1 Inhibitory Peptide 3cbz 
The Dvl2 PDZ Domain in Complex with the N2 Inhibitory Peptide 3cc0 
The Dvl2 PDZ Domain in Complex with the N3 Inhibitory Peptide 3ch8 
The crystal structure of PDZ-Fibronectin fusion protein 3cs0 
Crystal structure of DegP24 3cyy 
The crystal structure of ZO-1 PDZ2 in complex with the Cx43 peptide 3diw 
c-terminal beta-catenin bound TIP-1 structure 3dj1 
crystal structure of TIP-1 wild type 3dj3 
Crystal Structure of C-terminal Truncated TIP-1 (6-113) 3e17 
Crystal structure of the second PDZ domain from human Zona Occludens-2 3egg 
Crystal structure of a complex between Protein Phosphatase 1 alpha (PP1) and the PP1 binding and PDZ domains of Spinophilin 3egh 
Crystal structure of a complex between Protein Phosphatase 1 alpha (PP1), the PP1 binding and PDZ domains of Spinophilin and the small natural molecular toxin Nodularin-R 3fy5 
Dishevelled PDZ domain homodimer 3gcn 
Crystal structure of DegS H198P/D320A mutant modified by DFP in complex with OMP peptide (YQF) 3gco 
Crystal structure of DegS H198P/D320A mutant modified by DFP in complex with DNRDGNVYQF OMP peptide 3gds 
Crystal structure of DegS H198P/D320A mutant modified by DFP in complex with DNRDGNVYYF peptide 3gdu 
Crystal structure of DegS H198P/D320A mutant modified by DFP and in complex with YRF peptide 3gdv 
Crystal structure of DegS H198P/D320A mutant modified by DFP and in complex with YQF peptide 3gge 
Crystal structure of the PDZ domain of PDZ domain-containing protein GIPC2 3gj9 
crystal structure of TIP-1 in complex with c-terminal of Kir2.3 3gsl 
Crystal structure of PSD-95 tandem PDZ domains 1 and 2 3hpk 
Oxidized dimeric PICK1 PDZ in complex with the carboxyl tail peptide of GluR2 3hpm 
Oxidized dimeric PICK1 PDZ C46G mutant in complex with the carboxyl tail peptide of GluR2 3hvq 
Crystal structure of a complex between Protein Phosphatase 1 alpha (PP1) and the PP1 binding and PDZ domains of Neurabin 3i18 
Crystal Structure of the PDZ domain of the SdrC-like protein (Lmo2051) from Listeria monocytogenes, Northeast Structural Genomics Consortium Target LmR166B 3i1e 
Crystal Structure of the PDZ domain of the SdrC-like Protein (Lin2157) from Listeria innocua, Northeast Structural Genomics Consortium Target LkR136C 3i4w 
Crystal Structure of the third PDZ domain of PSD-95 3id2 
Crystal Structure of RseP PDZ2 domain 3id3 
Crystal Structure of RseP PDZ2 I304A domain 3id4 
Crystal Structure of RseP PDZ2 domain fused GKASPV peptide 3jxt 
Crystal structure of the third PDZ domain of SAP-102 in complex with a fluorogenic peptide-based ligand 3k1r 
Structure of harmonin NPDZ1 in complex with the SAM-PBM of Sans 3k50 
Crystal structure of Putative S41 protease (YP_211611.1) from Bacteroides fragilis NCTC 9343 at 2.00 A resolution 3k82 
Crystal Structure of the third PDZ domain of PSD-95 3khf 
The crystal structure of the PDZ domain of human Microtubule Associated Serine/Threonine Kinase 3 (MAST3) 3kzd 
Crystal Structure of Free T-cell Lymphoma Invasion and Metastasis-1 PDZ Domain 3kze 
Crystal Structure of T-cell Lymphoma Invasion and Metastasis-1 PDZ in Complex With SSRKEYYA Peptide 3l4f 
Crystal Structure of betaPIX Coiled-Coil Domain and Shank PDZ Complex 3lnx 
Second PDZ domain from human PTP1E 3lny 
Second PDZ domain from human PTP1E in complex with RA-GEF2 peptide 3mh4 
HtrA proteases are activated by a conserved mechanism that can be triggered by distinct molecular cues 3mh5 
HtrA proteases are activated by a conserved mechanism that can be triggered by distinct molecular cues 3mh6 
HtrA proteases are activated by a conserved mechanism that can be triggered by distinct molecular cues 3mh7 
HtrA proteases are activated by a conserved mechanism that can be triggered by distinct molecular cues 3nfk 
Crystal structure of the PTPN4 PDZ domain complexed with the C-terminus of a rabies virus G protein 3nfl 
Crystal structure of the PTPN4 PDZ domain complexed with the C-terminus of the GluN2A NMDA receptor subunit 3ngh 
Molecular Analysis of the Interaction of the HDL Receptor SR-BI with the Adaptor Protein PDZK1 3num 
Substrate induced remodeling of the active site regulates HtrA1 activity 3nzi 
Substrate induced remodeling of the active site regulates HtrA1 activity 3o46 
Crystal structure of the PDZ domain of MPP7 3o5n 
Tetrahydroquinoline carboxylates are potent inhibitors of the Shank PDZ domain, a putative target in autism disorders 3otp 
Crystal structure of the DegP dodecamer with a model substrate 3ou0 
re-refined 3CS0 3pdv 
Structure of the PDlim2 PDZ domain in complex with the C-terminal 6-peptide extension of NS1 3pdz 
SOLUTION STRUCTURE OF THE PDZ2 DOMAIN FROM HUMAN PHOSPHATASE HPTP1E 3ps4 
PDZ domain from Human microtubule-associated serine/threonine-protein kinase 1 3pv2 
Structure of Legionella fallonii DegQ (wt) 3pv3 
Structure of Legionella fallonii DegQ (S193A variant) 3pv4 
Structure of Legionella fallonii DegQ (Delta-PDZ2 variant) 3pv5 
Structure of Legionella fallonii DegQ (N189G/P190G variant) 3qdo 
Crystal Structure of PDZ domain of sorting nexin 27 (SNX27) fused to the Gly-Gly linker followed by C-terminal (ESESKV) of GIRK3 3qe1 
Crystal Structure of PDZ domain of sorting nexin 27 (SNX27) fused to the C-terminal residues (ESESKV) of GIRK3 3qgl 
Crystal Structure of PDZ domain of sorting nexin 27 (SNX27) in complex with the ESESKV peptide corresponding to the C-terminal tail of GIRK3 3qik 
Crystal structure of the first PDZ domain of PREX1 3qjm 
Structural flexibility of Shank PDZ domain is important for its binding to different ligands 3qjn 
Structural flexibility of Shank PDZ domain is important for its binding to different ligands 3qo6 
Crystal structure analysis of the plant protease Deg1 3r0h 
Structure of INAD PDZ45 in complex with NG2 peptide 3r68 
Molecular Analysis of the PDZ3 domain of PDZK1 3r69 
Molecular analysis of the interaction of the HDL-receptor SR-BI with the PDZ3 domain of its adaptor protein PDZK1 3rl7 
Crytal structure of hDLG1-PDZ1 complexed with APC 3rl8 
Crytal structure of hDLG1-PDZ2 complexed with APC 3rle 
Crystal Structure of GRASP55 GRASP domain (residues 7-208) 3sfj 
Crystal Structure of Tax-Interacting Protein-1 (TIP-1) PDZ domain bound to iCAL36 inhibitor peptide 3shu 
Crystal structure of ZO-1 PDZ3 3shw 
Crystal structure of ZO-1 PDZ3-SH3-Guk supramodule complex with Connexin-45 peptide 3soe 
Crystal Structure of the 3rd PDZ domain of the human Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 3 (MAGI3) 3stj 
Crystal structure of the protease + PDZ1 domain of DegQ from Escherichia coli 3suz 
Crystal structure of Rat Mint2 PPC 3tmh 
Crystal structure of dual-specific A-kinase anchoring protein 2 in complex with cAMP-dependent protein kinase A type II alpha and PDZK1 3tsv 
crystal structure of the third PDZ domain of the human ZO-1 MAGUK protein 3tsw 
crystal structure of the PDZ3-SH3-GUK core module of Human ZO-1 3tsz 
crystal structure of PDZ3-SH3-GUK core module from human ZO-1 in complex with 12mer peptide from human JAM-A cytoplasmic tail 3vqf 
Crystal Structure Analysis of the PDZ Domain Derived from the Tight Junction Regulating Protein 3vqg 
Crystal Structure Analysis of the PDZ Domain Derived from the Tight Junction Regulating Protein 3wkl 
The periplasmic PDZ tandem fragment of the RseP homologue from Aquifex aeolicus 3wkm 
The periplasmic PDZ tandem fragment of the RseP homologue from Aquifex aeolicus in complex with the Fab fragment 3zrt 
Crystal structure of human PSD-95 PDZ1-2 4a8a 
Asymmetric cryo-EM reconstruction of E. coli DegQ 12-mer in complex with lysozyme 4a8b 
Symmetrized cryo-EM reconstruction of E. coli DegQ 12-mer in complex with lysozymes 4a8c 
Symmetrized cryo-EM reconstruction of E. coli DegQ 12-mer in complex with a binding peptide 4a8d 
DegP dodecamer with bound OMP 4a9g 
Symmetrized cryo-EM reconstruction of E. coli DegQ 24-mer in complex with beta-casein 4amh 
Influence of circular permutation on the folding pathway of a PDZ domain 4c2c 
Crystal structure of the protease CtpB in an active state 4c2d 
Crystal structure of the protease CtpB in an active state 4c2e 
Crystal structure of the protease CtpB(S309A) present in a resting state 4c2f 
Crystal structure of the CtpB R168A mutant present in an active conformation 4c2g 
Crystal structure of CtpB(S309A) in complex with a peptide having a Val-Pro-Ala C-terminus 4c2h 
Crystal structure of the CtpB(V118Y) mutant 4cmz 
An intertwined homodimer of the PDZ homology domain of periaxin 4cn0 
An intertwined homodimer of the PDZ homology domain of AHNAK2 4e34 
Crystal structure of CFTR Associated Ligand (CAL) PDZ domain bound to iCAL36 (ANSRWPTSII) peptide 4e35 
Crystal structure of CFTR Associated Ligand (CAL) PDZ domain bound to iCAL36-L (ANSRWPTSIL) peptide 4e3b 
Crystal structure of Tax-Interacting Protein-1 (TIP-1) PDZ domain bound to iCAL36-L (ANSRWPTSIL) peptide 4edj 
Crystal structure of the GRASP55 GRASP Domain with a phosphomimetic mutation (S189D) 4f8k 
Molecular analysis of the interaction between the prostacyclin receptor and the first PDZ domain of PDZK1 4fgm 
Crystal structure of the aminopeptidase N family protein Q5QTY1 from Idiomarina loihiensis. Northeast Structural Genomics Consortium Target IlR60. 4g69 
Structure of the Human Discs Large 1 PDZ2 - Adenomatous Polyposis Coli Cytoskeletal Polarity Complex 4gvc 
Crystal Structure of T-cell Lymphoma Invasion and Metastasis-1 PDZ in complex with phosphorylated Syndecan1 Peptide 4gvd 
Crystal Structure of T-cell Lymphoma Invasion and Metastasis-1 PDZ in complex with Syndecan1 Peptide 4h11 
Interaction partners of PSD-93 studied by X-ray crystallography and fluorescent polarization spectroscopy 4hop 
Crystal structure of the computationally designed NNOS-Syntrophin complex 4ic6 
Crystal structure of Deg8 4jj0 
Crystal structure of MamP 4jj3 
Crystal structure of MamP in complex with iron(II) 4jl7 
Crystal Structure of the Chemokine Receptor CXCR2 in Complex with the First PDZ Domain of NHERF1 4joe 
CFTR Associated Ligand (CAL) PDZ domain bound to peptide A-iCAL36 (ANSRAPTSII) 4jof 
CFTR Associated Ligand (CAL) PDZ domain bound to peptide L-iCAL36 (ANSRLPTSII) 4jog 
CFTR Associated Ligand (CAL) PDZ domain bound to peptide V-iCAL36 (ANSRVPTSII) 4joh 
CFTR Associated Ligand (CAL) PDZ domain bound to peptide H-iCAL36 (ANSRHPTSII) 4joj 
CFTR Associated Ligand (CAL) domain bound to peptide F-iCAL36 (ANSRFPTSII) 4jok 
CFTR Associated Ligand (CAL) PDZ domain bound to peptide Y-iCAL36 (ANSRYPTSII) 4jop 
CFTR Associated Ligand (CAL) PDZ bound to HPV16 E6 oncoprotein C-terminal peptide (TRRETQL) 4jor 
CFTR Associated Ligand (CAL) PDZ domain bound to HPV18 E6 oncoprotein C-terminal peptide (RLQRRRETQV) 4k6y 
CFTR Associated Ligand (CAL) PDZ domain bound to peptide iCAL36-Q (ANSRWQTSII) 4k72 
CFTR Associated Ligand (CAL) PDZ domain bound to peptide iCAL36-VQD (ANSRVQDSII) 4k75 
CFTR Associated Ligand (CAL) PDZ domain bound to peptide iCAL36-QDTRL (ANSRWQDTRL) 4k76 
CFTR Associated Ligand (CAL) PDZ domain bound to peptide iCAL36-TRL (ANSRWPTTRL) 4k78 
CFTR Associated Ligand (CAL) E317A PDZ domain bound to peptide iCAL36-QDTRL (ANSRWQDTRL) 4kfw 
Structural insight into Golgi membrane stacking by GRASP65 and GRASP55 4l8n 
Crystal structure of a PDZ domain protein (BDI_1242) from Parabacteroides distasonis ATCC 8503 at 2.50 A resolution 4lmm 
Crystal structure of NHERF1 PDZ1 domain complexed with the CXCR2 C-terminal tail in P21 space group 4mpa 
Crystal structure of NHERF1-CXCR2 signaling complex in P21 space group 4n6x 
Crystal Structure of the Chemokine Receptor CXCR2 in Complex with the First PDZ Domain of NHERF1 4nmo 
4NMO 4nmp 
4NMP 4nmq 
4NMQ 4nmr 
4NMR 4nms 
4NMS 4nmt 
4NMT 4nmv 
4NMV 4nnl 
4NNL 4nnm 
4NNM 4nxp 
4NXP 4nxq 
4NXQ 4nxr 
4NXR 4o06 
1.15A Resolution Structure of the Proteasome Assembly Chaperone Nas2 PDZ Domain 4oaj 
Crystal structure of the complex between SAP97 PDZ2 and 5HT2A receptor peptide 4oeo 
4OEO 4oep 
4OEP 4p0c 
Crystal Structure of NHERF2 PDZ1 Domain in Complex with LPA2 4p2a 
4P2A 4pqw 
Crystal Structure of Phospholipase C beta 3 in Complex with PDZ1 of NHERF1 4q2n 
4Q2N 4q2o 
4Q2O 4q2p 
4Q2P 4q2q 
4Q2Q 4q3h 
The crystal structure of NHERF1 PDZ2 CXCR2 complex revealed by the NHERF1 CXCR2 chimeric protein 4q6h 
CFTR Associated Ligand (CAL) bound to last 6 residues of CFTR (decameric peptide: iCAL36VQDTRL) 4q6s 
4Q6S 4ql6 
4QL6 4r2z 
4R2Z 4ri0 
4RI0 4rqy 
4RQY 4rqz 
4RQZ 4rr0 
4RR0 4rr1 
4RR1 4uu5 
4UU5 4uu6 
4UU6 4wsi 
4WSI 4wyt 
4WYT 4wyu 
4WYU 4xh7 
4XH7 4xhv 
4XHV 4ydp 
4YDP 4ynn 
4YNN 4yo1 
4YO1 4yyx 
4YYX 4z33 
4Z33 4z8j 
4Z8J 5a2p 
5A2P 5b6l 
5B6L 5d13 
5D13 5dth 
5DTH 5e6p 
5E6P 5elq 
5ELQ 5em9 
5EM9 5ema 
5EMA 5emb 
5EMB 5eyz 
5EYZ 5ez0 
5EZ0 5f3x 
5F3X 5f67 
5F67 5fb8 
5FB8 5fht 
5FHT 5g1d 
5G1D 5g1e 
5G1E 5glj 
5GLJ 5gnd 
5GND 5heb 
5HEB 5hed 
5HED 5het 
5HET 5hey 
5HEY 5hf1 
5HF1 5hfb 
5HFB 5hfc 
5HFC 5hff 
5HFF 5hj1 
5HJ1 5i7z 
5I7Z 5izu 
5IZU 5jd8 
5JD8 5jxb 
5JXB - Links (links to other resources describing this domain)
-
INTERPRO IPR001478 PFAM PF00595

