The domain within your query sequence starts at position 108 and ends at position 179; the E-value for the S4 domain shown below is 6.84e-4.
RRLQTQVFKLGLAKSIHHARVLIRQRHIRVRKQVVNIPSFIVRLDSQKHIDFSLRSPYGG GRPGRVKRKNAK
S4S4 RNA-binding domain |
|---|
| SMART accession number: | SM00363 |
|---|---|
| Description: | - |
| Interpro abstract (IPR002942): | The S4 domain is a small domain consisting of 60-65 amino acid residues that was detected in the bacterial ribosomal protein S4, eukaryotic ribosomal S9, two families of pseudouridine synthases, a novel family of predicted RNA methylases, a yeast protein containing a pseudouridine synthetase and a deaminase domain, bacterial tyrosyl-tRNA synthetases, and a number of uncharacterised, small proteins that may be involved in translation regulation [ (PUBMED:10093218) ]. The S4 domain probably mediates binding to RNA [ (PUBMED:9707415) ]. |
| GO function: | RNA binding (GO:0003723) |
| Family alignment: |
There are 146383 S4 domains in 146322 proteins in SMART's nrdb database.
Click on the following links for more information.
- Evolution (species in which this domain is found)
-
Taxonomic distribution of proteins containing S4 domain.
This tree includes only several representative species. The complete taxonomic breakdown of all proteins with S4 domain is also avaliable.
Click on the protein counts, or double click on taxonomic names to display all proteins containing S4 domain in the selected taxonomic class.
- Cellular role (predicted cellular role)
-
Binding / catalysis: RNA-binding
- Literature (relevant references for this domain)
-
Primary literature is listed below; Automatically-derived, secondary literature is also avaliable.
- Aravind L, Koonin EV
- Novel predicted RNA-binding domains associated with the translation machinery.
- J Mol Evol. 1999; 48: 291-302
- Display abstract
Two previously undetected domains were identified in a variety of RNA-binding proteins, particularly RNA-modifying enzymes, using methods for sequence profile analysis. A small domain consisting of 60-65 amino acid residues was detected in the ribosomal protein S4, two families of pseudouridine synthases, a novel family of predicted RNA methylases, a yeast protein containing a pseudouridine synthetase and a deaminase domain, bacterial tyrosyl-tRNA synthetases, and a number of uncharacterized, small proteins that may be involved in translation regulation. Another novel domain, designated PUA domain, after PseudoUridine synthase and Archaeosine transglycosylase, was detected in archaeal and eukaryotic pseudouridine synthases, archaeal archaeosine synthases, a family of predicted ATPases that may be involved in RNA modification, a family of predicted archaeal and bacterial rRNA methylases. Additionally, the PUA domain was detected in a family of eukaryotic proteins that also contain a domain homologous to the translation initiation factor eIF1/SUI1; these proteins may comprise a novel type of translation factors. Unexpectedly, the PUA domain was detected also in bacterial and yeast glutamate kinases; this is compatible with the demonstrated role of these enzymes in the regulation of the expression of other genes. We propose that the S4 domain and the PUA domain bind RNA molecules with complex folded structures, adding to the growing collection of nucleic acid-binding domains associated with DNA and RNA modification enzymes. The evolution of the translation machinery components containing the S4, PUA, and SUI1 domains must have included several events of lateral gene transfer and gene loss as well as lineage-specific domain fusions.
- Markus MA, Gerstner RB, Draper DE, Torchia DA
- The solution structure of ribosomal protein S4 delta41 reveals two subdomains and a positively charged surface that may interact with RNA.
- EMBO J. 1998; 17: 4559-71
- Display abstract
S4 is one of the first proteins to bind to 16S RNA during assembly of the prokaryotic ribosome. Residues 43-200 of S4 from Bacillus stearothermophilus (S4 Delta41) bind specifically to both 16S rRNA and to a pseudoknot within the alpha operon mRNA. As a first step toward understanding how S4 recognizes and organizes RNA, we have solved the structure of S4 Delta41 in solution by multidimensional heteronuclear nuclear magnetic resonance spectroscopy. The fold consists of two globular subdomains, one comprised of four helices and the other comprised of a five-stranded antiparallel beta-sheet and three helices. Although cross-linking studies suggest that residues between helices alpha2 and alpha3 are close to RNA, the concentration of positive charge along the crevice between the two subdomains suggests that this could be an RNA-binding site. In contrast to the L11 RNA-binding domain studied previously, S4 Delta41 shows no fast local motions, suggesting that it has less capacity for refolding to fit RNA. The independently determined crystal structure of S4 Delta41 shows similar features, although there is small rotation of the subdomains compared with the solution structure. The relative orientation of the subdomains in solution will be verified with further study.
- Metabolism (metabolic pathways involving proteins which contain this domain)
-
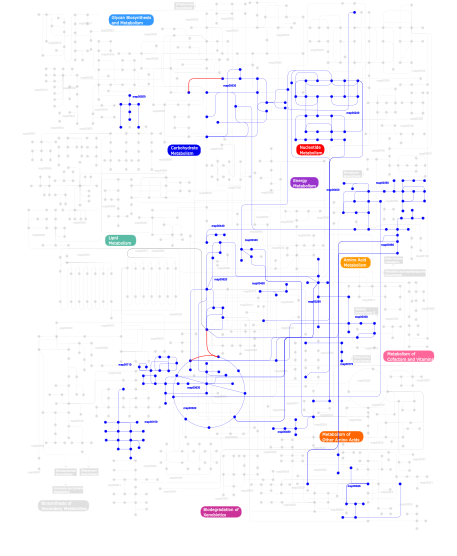
Click the image to view the interactive version of the map in iPath% proteins involved KEGG pathway ID Description 32.37 map03010 Ribosome 28.94  map00400
map00400Phenylalanine, tyrosine and tryptophan biosynthesis 28.87 map00970 Aminoacyl-tRNA biosynthesis 6.78  map00240
map00240Pyrimidine metabolism 0.30  map00350
map00350Tyrosine metabolism 0.30  map00450
map00450Selenoamino acid metabolism 0.30  map00340
map00340Histidine metabolism 0.30  map00380
map00380Tryptophan metabolism 0.30  map00440
map00440Aminophosphonate metabolism 0.30  map00626
map00626Naphthalene and anthracene degradation 0.30  map00150
map00150Androgen and estrogen metabolism 0.15  map00260
map00260Glycine, serine and threonine metabolism 0.15  map00300
map00300Lysine biosynthesis 0.08  map00770
map00770Pantothenate and CoA biosynthesis 0.08  map00620
map00620Pyruvate metabolism 0.08  map00630
map00630Glyoxylate and dicarboxylate metabolism 0.08  map00020
map00020Citrate cycle (TCA cycle) 0.08  map00480
map00480Glutathione metabolism 0.08  map00710
map00710Carbon fixation 0.08  map00550
map00550Peptidoglycan biosynthesis 0.08  map00030
map00030Pentose phosphate pathway This information is based on mapping of SMART genomic protein database to KEGG orthologous groups. Percentage points are related to the number of proteins with S4 domain which could be assigned to a KEGG orthologous group, and not all proteins containing S4 domain. Please note that proteins can be included in multiple pathways, ie. the numbers above will not always add up to 100%.
- Structure (3D structures containing this domain)
3D Structures of S4 domains in PDB
PDB code Main view Title 1c05 
SOLUTION STRUCTURE OF RIBOSOMAL PROTEIN S4 DELTA 41, REFINED WITH DIPOLAR COUPLINGS (MINIMIZED AVERAGE STRUCTURE) 1c06 
SOLUTION STRUCTURE OF RIBOSOMAL PROTEIN S4 DELTA 41, REFINED WITH DIPOLAR COUPLINGS (ENSEMBLE OF 16 STRUCTURES) 1dm9 
HEAT SHOCK PROTEIN 15 KD 1eg0 
FITTING OF COMPONENTS WITH KNOWN STRUCTURE INTO AN 11.5 A CRYO-EM MAP OF THE E.COLI 70S RIBOSOME 1fjg 
STRUCTURE OF THE THERMUS THERMOPHILUS 30S RIBOSOMAL SUBUNIT IN COMPLEX WITH THE ANTIBIOTICS STREPTOMYCIN, SPECTINOMYCIN, AND PAROMOMYCIN 1fka 
STRUCTURE OF FUNCTIONALLY ACTIVATED SMALL RIBOSOMAL SUBUNIT AT 3.3 A RESOLUTION 1h3e 
Tyrosyl-tRNA synthetase from Thermus thermophilus complexed with wild-type tRNAtyr(GUA) and with ATP and tyrosinol 1h3f 
Tyrosyl-tRNA synthetase from Thermus thermophilus complexed tyrosinol 1hnw 
STRUCTURE OF THE THERMUS THERMOPHILUS 30S RIBOSOMAL SUBUNIT IN COMPLEX WITH TETRACYCLINE 1hnx 
STRUCTURE OF THE THERMUS THERMOPHILUS 30S RIBOSOMAL SUBUNIT IN COMPLEX WITH PACTAMYCIN 1hnz 
STRUCTURE OF THE THERMUS THERMOPHILUS 30S RIBOSOMAL SUBUNIT IN COMPLEX WITH HYGROMYCIN B 1hr0 
CRYSTAL STRUCTURE OF INITIATION FACTOR IF1 BOUND TO THE 30S RIBOSOMAL SUBUNIT 1i94 
CRYSTAL STRUCTURES OF THE SMALL RIBOSOMAL SUBUNIT WITH TETRACYCLINE, EDEINE AND IF3 1i95 
CRYSTAL STRUCTURE OF THE 30S RIBOSOMAL SUBUNIT FROM THERMUS THERMOPHILUS IN COMPLEX WITH EDEINE 1i96 
CRYSTAL STRUCTURE OF THE 30S RIBOSOMAL SUBUNIT FROM THERMUS THERMOPHILUS IN COMPLEX WITH THE TRANSLATION INITIATION FACTOR IF3 (C-TERMINAL DOMAIN) 1i97 
CRYSTAL STRUCTURE OF THE 30S RIBOSOMAL SUBUNIT FROM THERMUS THERMOPHILUS IN COMPLEX WITH TETRACYCLINE 1ibk 
STRUCTURE OF THE THERMUS THERMOPHILUS 30S RIBOSOMAL SUBUNIT IN COMPLEX WITH THE ANTIBIOTIC PAROMOMYCIN 1ibl 
STRUCTURE OF THE THERMUS THERMOPHILUS 30S RIBOSOMAL SUBUNIT IN COMPLEX WITH A MESSENGER RNA FRAGMENT AND COGNATE TRANSFER RNA ANTICODON STEM-LOOP BOUND AT THE A SITE AND WITH THE ANTIBIOTIC PAROMOMYCIN 1ibm 
STRUCTURE OF THE THERMUS THERMOPHILUS 30S RIBOSOMAL SUBUNIT IN COMPLEX WITH A MESSENGER RNA FRAGMENT AND COGNATE TRANSFER RNA ANTICODON STEM-LOOP BOUND AT THE A SITE 1j5e 
Structure of the Thermus thermophilus 30S Ribosomal Subunit 1jgo 
The Path of Messenger RNA Through the Ribosome. THIS FILE, 1JGO, CONTAINS THE 30S RIBOSOME SUBUNIT, THREE TRNA, AND MRNA MOLECULES. 50S RIBOSOME SUBUNIT IS IN THE FILE 1GIY 1jgp 
The Path of Messenger RNA Through the Ribosome. THIS FILE, 1JGP, CONTAINS THE 30S RIBOSOME SUBUNIT, THREE TRNA, AND MRNA MOLECULES. 50S RIBOSOME SUBUNIT IS IN THE FILE 1GIY 1jgq 
The Path of Messenger RNA Through the Ribosome. THIS FILE, 1JGQ, CONTAINS THE 30S RIBOSOME SUBUNIT, THREE TRNA, AND MRNA MOLECULES. 50S RIBOSOME SUBUNIT IS IN THE FILE 1GIY 1jh3 
Solution structure of tyrosyl-tRNA synthetase C-terminal domain. 1jii 
Crystal structure of S. aureus TyrRS in complex with SB-219383 1jij 
Crystal structure of S. aureus TyrRS in complex with SB-239629 1jik 
Crystal structure of S. aureus TyrRS in complex with SB-243545 1jil 
Crystal structure of S. aureus TyrRS in complex with SB284485 1ksk 
STRUCTURE OF RSUA 1ksl 
STRUCTURE OF RSUA 1ksv 
STRUCTURE OF RSUA 1ml5 
Structure of the E. coli ribosomal termination complex with release factor 2 1n32 
Structure of the Thermus thermophilus 30S ribosomal subunit bound to codon and near-cognate transfer RNA anticodon stem-loop mismatched at the first codon position at the a site with paromomycin 1n33 
Structure of the Thermus thermophilus 30S ribosomal subunit bound to codon and near-cognate transfer rna anticodon stem-loop mismatched at the second codon position at the a site with paromomycin 1n34 
Structure of the Thermus thermophilus 30S ribosomal subunit in the presence of codon and crystallographically disordered near-cognate transfer rna anticodon stem-loop mismatched at the first codon position 1n36 
Structure of the Thermus thermophilus 30S ribosomal subunit in the presence of crystallographically disordered codon and near-cognate transfer RNA anticodon stem-loop mismatched at the second codon position 1qd7 
PARTIAL MODEL FOR 30S RIBOSOMAL SUBUNIT 1qyu 
Structure of the catalytic domain of 23S rRNA pseudouridine synthase RluD 1v9f 
Crystal structure of catalytic domain of pseudouridine synthase RluD from Escherichia coli 1vio 
Crystal structure of pseudouridylate synthase 1vvj 
1VVJ 1vy4 
1VY4 1vy5 
1VY5 1vy6 
1VY6 1vy7 
1VY7 1xmo 
Crystal Structure of mnm5U34t6A37-tRNALysUUU Complexed with AAG-mRNA in the Decoding Center 1xmq 
Crystal Structure of t6A37-ASLLysUUU AAA-mRNA Bound to the Decoding Center 1xnq 
Structure of an Inosine-Adenine Wobble Base Pair Complex in the Context of the Decoding Center 1xnr 
Crystal Structure of an Inosine-Cytosine Wobble Base Pair in the Context of the Decoding Center 2cqj 
Solution structure of the S4 domain of U3 small nucleolar ribonucleoprotein protein IMP3 homolog 2e5l 
A snapshot of the 30S ribosomal subunit capturing mRNA via the Shine- Dalgarno interaction 2f4v 
30S ribosome + designer antibiotic 2hhh 
Crystal structure of kasugamycin bound to the 30S ribosomal subunit 2ist 
crystal structure of RluD from E. coli 2jan 
TYROSYL-TRNA SYNTHETASE FROM MYCOBACTERIUM TUBERCULOSIS IN UNLIGANDED STATE 2k6p 
Solution Structure of hypothetical protein, HP1423 2ts1 
STRUCTURE OF TYROSYL-T/RNA SYNTHETASE REFINED AT 2.3 ANGSTROMS RESOLUTION. INTERACTION OF THE ENZYME WITH THE TYROSYL ADENYLATE INTERMEDIATE 2uu9 
Structure of the Thermus thermophilus 30S ribosomal subunit complexed with a Valine-ASL with cmo5U in position 34 bound to an mRNA with a GUG-codon in the A-site and paromomycin. 2uua 
Structure of the Thermus thermophilus 30S ribosomal subunit complexed with a Valine-ASL with cmo5U in position 34 bound to an mRNA with a GUC-codon in the A-site and paromomycin. 2uub 
Structure of the Thermus thermophilus 30S ribosomal subunit complexed with a Valine-ASL with cmo5U in position 34 bound to an mRNA with a GUU-codon in the A-site and paromomycin. 2uuc 
Structure of the Thermus thermophilus 30S ribosomal subunit complexed with a Valine-ASL with cmo5U in position 34 bound to an mRNA with a GUA-codon in the A-site and paromomycin. 2uxb 
Crystal structure of an extended tRNA anticodon stem loop in complex with its cognate mRNA GGGU in the context of the Thermus thermophilus 30S subunit. 2uxc 
Crystal structure of an extended tRNA anticodon stem loop in complex with its cognate mRNA UCGU in the context of the Thermus thermophilus 30S subunit. 2uxd 
Crystal structure of an extended tRNA anticodon stem loop in complex with its cognate mRNA CGGG in the context of the Thermus thermophilus 30S subunit. 2vqe 
Modified uridines with C5-methylene substituents at the first position of the tRNA anticodon stabilize U-G wobble pairing during decoding 2vqf 
Modified uridines with C5-methylene substituents at the first position of the tRNA anticodon stabilize U-G wobble pairing during decoding 2ykr 
30S ribosomal subunit with RsgA bound in the presence of GMPPNP 2zm6 
Crystal structure of the Thermus thermophilus 30S ribosomal subunit 3bbu 
The Hsp15 protein fitted into the low resolution Cryo-EM map of the 50S.nc-tRNA.Hsp15 complex 3dh3 
Crystal Structure of RluF in complex with a 22 nucleotide RNA substrate 3hp7 
Putative hemolysin from Streptococcus thermophilus. 3j6x 
3J6X 3j6y 
3J6Y 3j77 
3J77 3j78 
3J78 3j7a 
3J7A 3j7p 
3J7P 3j7r 
3J7R 3j80 
3J80 3j81 
3J81 3j9w 
3J9W 3j9y 
3J9Y 3j9z 
3J9Z 3ja1 
3JA1 3jag 
3JAG 3jah 
3JAH 3jai 
3JAI 3jaj 
3JAJ 3jam 
3JAM 3jan 
3JAN 3jap 
3JAP 3jaq 
3JAQ 3jbn 
3JBN 3jbo 
3JBO 3jbp 
3JBP 3jbu 
3JBU 3jbv 
3JBV 3jcd 
3JCD 3jce 
3JCE 3jcj 
3JCJ 3jcn 
3JCN 3kbg 
Crystal structure of the 30S ribosomal protein S4e from Thermoplasma acidophilum. Northeast Structural Genomics Consortium Target TaR28. 3oto 
Crystal Structure of the 30S ribosomal subunit from a KsgA mutant of Thermus thermophilus (HB8) 3t1h 
Structure of the Thermus thermophilus 30S ribosomal subunit complexed with a human anti-codon stem loop (HASL) of transfer RNA lysine 3 (tRNALys3) bound to an mRNA with an AAA-codon in the A-site and Paromomycin 3t1y 
Structure of the Thermus thermophilus 30S ribosomal subunit complexed with a human anti-codon stem loop (HASL) of transfer RNA Lysine 3 (TRNALYS3) bound to an mRNA with an AAG-codon in the A-site and paromomycin 3ts1 
STRUCTURE OF TYROSYL-T/RNA SYNTHETASE REFINED AT 2.3 ANGSTROMS RESOLUTION. INTERACTION OF THE ENZYME WITH THE TYROSYL ADENYLATE INTERMEDIATE 4a2i 
Cryo-electron Microscopy Structure of the 30S Subunit in Complex with the YjeQ Biogenesis Factor 4adv 
Structure of the E. coli methyltransferase KsgA bound to the E. coli 30S ribosomal subunit 4aqy 
Structure of ribsome-apramycin complexes 4b3m 
Crystal structure of the 30S ribosome in complex with compound 1 4b3r 
Crystal structure of the 30S ribosome in complex with compound 30 4b3s 
Crystal structure of the 30S ribosome in complex with compound 37 4b3t 
Crystal structure of the 30S ribosome in complex with compound 39 4bts 
4BTS 4d5l 
4D5L 4d61 
4D61 4dr1 
Crystal structure of the apo 30S ribosomal subunit from Thermus thermophilus (HB8) 4dr2 
Crystal structure of the Thermus thermophilus (HB8) 30S ribosomal subunit with multiple copies of paromomycin molecules bound 4dr3 
Crystal structure of the Thermus thermophilus (HB8) 30S ribosomal subunit with streptomycin bound 4dr4 
Crystal structure of the Thermus thermophilus (HB8) 30S ribosomal subunit with codon, cognate transfer RNA anticodon stem-loop and multiple copies of paromomycin molecules bound 4dr5 
Crystal structure of the Thermus thermophilus (HB8) 30S ribosomal subunit with codon, crystallographically disordered cognate transfer RNA anticodon stem-loop and streptomycin bound 4dr6 
Crystal structure of the Thermus thermophilus (HB8) 30S ribosomal subunit with codon, near-cognate transfer RNA anticodon stem-loop mismatched at the first codon position and streptomycin bound 4dr7 
Crystal structure of the Thermus thermophilus (HB8) 30S ribosomal subunit with codon, crystallographically disordered near-cognate transfer RNA anticodon stem-loop mismatched at the second codon position, and streptomycin bound 4duy 
Crystal structure of the Thermus thermophilus 30S ribosomal subunit with a 16S rRNA mutation, U13C 4duz 
Crystal structure of the Thermus thermophilus 30S ribosomal subunit with a 16S rRNA mutation, U13C, bound with streptomycin 4dv0 
Crystal structure of the Thermus thermophilus 30S ribosomal subunit with a 16S rRNA mutation, U20G 4dv1 
Crystal structure of the Thermus thermophilus 30S ribosomal subunit with a 16S rRNA mutation, U20G, bound with streptomycin 4dv2 
Crystal structure of the Thermus thermophilus 30S ribosomal subunit with a 16S rRNA mutation, C912A 4dv3 
Crystal structure of the Thermus thermophilus 30S ribosomal subunit with a 16S rRNA mutation, C912A, bound with streptomycin 4dv4 
Crystal structure of the Thermus thermophilus 30S ribosomal subunit with a 16S rRNA mutation, A914G 4dv5 
Crystal structure of the Thermus thermophilus 30S ribosomal subunit with a 16S rRNA mutation, A914G, bound with streptomycin 4dv6 
Crystal structure of the Thermus thermophilus 30S ribosomal subunit with a 16S rRNA mutation, A915G 4dv7 
Crystal structure of the Thermus thermophilus 30S ribosomal subunit with a 16S rRNA mutation, A915G, bound with streptomycin 4gkj 
Structure of the Thermus thermophilus 30S ribosomal subunit complexed with a human mitochondrial anticodon stem loop (ASL) of transfer RNA Methionine (TRNAMET) bound to an mRNA with an AUG-codon in the A-site and paromomycin. 4gkk 
Structure of the Thermus thermophilus 30S ribosomal subunit complexed with a human mitochondrial anticodon stem loop (ASL) of transfer RNA Methionine (TRNAMET) bound to an mRNA with an AUA-codon in the A-site and paromomycin 4ji0 
Crystal Structure of 30S ribosomal subunit from Thermus thermophilus 4ji1 
Crystal Structure of 30S ribosomal subunit from Thermus thermophilus 4ji2 
Crystal Structure of 30S ribosomal subunit from Thermus thermophilus 4ji3 
Crystal Structure of 30S ribosomal subunit from Thermus thermophilus 4ji4 
Crystal Structure of 30S ribosomal subunit from Thermus thermophilus 4ji5 
Crystal Structure of 30S ribosomal subunit from Thermus thermophilus 4ji6 
Crystal Structure of 30S ribosomal subunit from Thermus thermophilus 4ji7 
Crystal Structure of 30S ribosomal subunit from Thermus thermophilus 4ji8 
Crystal Structure of 30S ribosomal subunit from Thermus thermophilus 4jv5 
Crystal structures of pseudouridinilated stop codons with ASLs 4jya 
Crystal structures of pseudouridinilated stop codons with ASLs 4k0k 
Crystal structure of the Thermus thermophilus 30S ribosomal subunit complexed with a serine-ASL and mRNA containing a stop codon 4khp 
Structure of the Thermus thermophilus 30S ribosomal subunit in complex with de-6-MSA-pactamycin 4kvb 
4KVB 4kzx 
Rabbit 40S ribosomal subunit in complex with eIF1. 4kzy 
Rabbit 40S ribosomal subunit in complex with eIF1 and eIF1A. 4kzz 
Rabbit 40S ribosomal subunit in complex with mRNA, initiator tRNA and eIF1A 4l47 
4L47 4l71 
4L71 4lab 
Crystal structure of the catalytic domain of RluB 4lel 
4LEL 4lf4 
4LF4 4lf5 
4LF5 4lf6 
4LF6 4lf7 
4LF7 4lf8 
4LF8 4lf9 
4LF9 4lfa 
4LFA 4lfb 
4LFB 4lfc 
4LFC 4lfz 
4LFZ 4lgt 
Crystal structure of the catalytic domain of RluB in complex with a 21-nucleotide RNA substrate 4lnt 
4LNT 4lsk 
4LSK 4lt8 
4LT8 4nxm 
Crystal Structure of the 30S ribosomal subunit from a GidB (RsmG) mutant of Thermus thermophilus (HB8) 4nxn 
Crystal Structure of the 30S ribosomal subunit from a GidB (RsmG) mutant of Thermus thermophilus (HB8), bound with streptomycin 4oud 
4OUD 4ox9 
Crystal structure of the aminoglycoside resistance methyltransferase NpmA bound to the 30S ribosomal subunit 4p6f 
4P6F 4p70 
4P70 4tua 
4TUA 4tub 
4TUB 4tuc 
4TUC 4tud 
4TUD 4tue 
4TUE 4u1u 
4U1U 4u1v 
4U1V 4u20 
4U20 4u24 
4U24 4u25 
4U25 4u26 
4U26 4u27 
4U27 4u3m 
4U3M 4u3n 
4U3N 4u3u 
4U3U 4u4n 
4U4N 4u4o 
4U4O 4u4q 
4U4Q 4u4r 
4U4R 4u4u 
4U4U 4u4y 
4U4Y 4u4z 
4U4Z 4u50 
4U50 4u51 
4U51 4u52 
4U52 4u53 
4U53 4u55 
4U55 4u56 
4U56 4u6f 
4U6F 4uer 
4UER 4ug0 
4UG0 4ujc 
4UJC 4ujd 
4UJD 4uje 
4UJE 4v3p 
4V3P 4v42 
4V42 4v47 
4V47 4v48 
4V48 4v49 
4V49 4v4a 
4V4A 4v4b 
4V4B 4v4g 
4V4G 4v4h 
4V4H 4v4i 
4V4I 4v4j 
4V4J 4v4n 
4V4N 4v4p 
4V4P 4v4q 
4V4Q 4v4r 
4V4R 4v4s 
4V4S 4v4t 
4V4T 4v4v 
4V4V 4v4w 
4V4W 4v4x 
4V4X 4v4y 
4V4Y 4v4z 
4V4Z 4v50 
4V50 4v51 
4V51 4v52 
4V52 4v53 
4V53 4v54 
4V54 4v55 
4V55 4v56 
4V56 4v57 
4V57 4v5a 
4V5A 4v5b 
4V5B 4v5c 
4V5C 4v5d 
4V5D 4v5e 
4V5E 4v5f 
4V5F 4v5g 
4V5G 4v5h 
4V5H 4v5j 
4V5J 4v5k 
4V5K 4v5l 
4V5L 4v5m 
4V5M 4v5n 
4V5N 4v5o 
4V5O 4v5p 
4V5P 4v5q 
4V5Q 4v5r 
4V5R 4v5s 
4V5S 4v5y 
4V5Y 4v5z 
4V5Z 4v61 
4V61 4v63 
4V63 4v64 
4V64 4v65 
4V65 4v66 
4V66 4v67 
4V67 4v68 
4V68 4v69 
4V69 4v6a 
4V6A 4v6c 
4V6C 4v6d 
4V6D 4v6e 
4V6E 4v6f 
4V6F 4v6g 
4V6G 4v6i 
4V6I 4v6k 
4V6K 4v6l 
4V6L 4v6m 
4V6M 4v6n 
4V6N 4v6o 
4V6O 4v6p 
4V6P 4v6q 
4V6Q 4v6r 
4V6R 4v6s 
4V6S 4v6t 
4V6T 4v6u 
4V6U 4v6v 
4V6V 4v6w 
4V6W 4v6x 
4V6X 4v6y 
4V6Y 4v6z 
4V6Z 4v70 
4V70 4v71 
4V71 4v72 
4V72 4v73 
4V73 4v74 
4V74 4v75 
4V75 4v76 
4V76 4v77 
4V77 4v78 
4V78 4v79 
4V79 4v7a 
4V7A 4v7b 
4V7B 4v7c 
4V7C 4v7d 
4V7D 4v7e 
4V7E 4v7h 
4V7H 4v7i 
4V7I 4v7j 
4V7J 4v7k 
4V7K 4v7l 
4V7L 4v7m 
4V7M 4v7p 
4V7P 4v7r 
4V7R 4v7s 
4V7S 4v7t 
4V7T 4v7u 
4V7U 4v7v 
4V7V 4v7w 
4V7W 4v7x 
4V7X 4v7y 
4V7Y 4v7z 
4V7Z 4v83 
4V83 4v84 
4V84 4v85 
4V85 4v87 
4V87 4v88 
4V88 4v89 
4V89 4v8a 
4V8A 4v8b 
4V8B 4v8c 
4V8C 4v8d 
4V8D 4v8e 
4V8E 4v8f 
4V8F 4v8g 
4V8G 4v8h 
4V8H 4v8i 
4V8I 4v8j 
4V8J 4v8m 
4V8M 4v8n 
4V8N 4v8o 
4V8O 4v8q 
4V8Q 4v8u 
4V8U 4v8x 
4V8X 4v8y 
4V8Y 4v8z 
4V8Z 4v90 
4V90 4v92 
4V92 4v95 
4V95 4v97 
4V97 4v9a 
4V9A 4v9b 
4V9B 4v9c 
4V9C 4v9d 
4V9D 4v9h 
4V9H 4v9i 
4V9I 4v9j 
4V9J 4v9k 
4V9K 4v9l 
4V9L 4v9m 
4V9M 4v9n 
4V9N 4v9o 
4V9O 4v9p 
4V9P 4v9q 
4V9Q 4v9r 
4V9R 4v9s 
4V9S 4w29 
4W29 4w2e 
4W2E 4w2f 
4W2F 4w2g 
4W2G 4w2h 
4W2H 4w2i 
4W2I 4w4g 
4W4G 4wf1 
4WF1 4woi 
4WOI 4wpo 
4WPO 4wq1 
4WQ1 4wqf 
4WQF 4wqr 
4WQR 4wqu 
4WQU 4wqy 
4WQY 4wr6 
4WR6 4wra 
4WRA 4wro 
4WRO 4wsd 
4WSD 4wsm 
4WSM 4wt1 
4WT1 4wt8 
4WT8 4wu1 
4WU1 4www 
4WWW 4wzd 
4WZD 4wzo 
4WZO 4x62 
4X62 4x64 
4X64 4x65 
4X65 4x66 
4X66 4xej 
4XEJ 4y4o 
4Y4O 4y4p 
4Y4P 4ybb 
4YBB 4yhh 
4YHH 4ypb 
4YPB 4yy3 
4YY3 4yzv 
4YZV 4z3s 
4Z3S 4z8c 
4Z8C 4zer 
4ZER 4zsn 
4ZSN 5a2q 
5A2Q 5a9z 
5A9Z 5aa0 
5AA0 5afi 
5AFI 5aj0 
5AJ0 5br8 
5BR8 5czp 
5CZP 5d8b 
5D8B 5dat 
5DAT 5dc3 
5DC3 5dfe 
5DFE 5dox 
5DOX 5doy 
5DOY 5e7k 
5E7K 5e81 
5E81 5el4 
5EL4 5el5 
5EL5 5el6 
5EL6 5el7 
5EL7 5f8k 
5F8K 5fci 
5FCI 5fcj 
5FCJ 5fdu 
5FDU 5fdv 
5FDV 5flx 
5FLX 5hau 
5HAU 5hcp 
5HCP 5hcq 
5HCQ 5hcr 
5HCR 5hd1 
5HD1 5i4l 
5I4L 5ib7 
5IB7 5ib8 
5IB8 5ibb 
5IBB 5imq 
5IMQ 5imr 
5IMR 5iqr 
5IQR 5it7 
5IT7 5it8 
5IT8 5it9 
5IT9 5iwa 
5IWA 5j30 
5J30 5j3c 
5J3C 5j4b 
5J4B 5j4c 
5J4C 5j4d 
5J4D 5j5b 
5J5B 5j7l 
5J7L 5j88 
5J88 5j8a 
5J8A 5j8b 
5J8B 5j91 
5J91 5jb3 
5JB3 5jc9 
5JC9 5jpq 
5JPQ 5jte 
5JTE 5ju8 
5JU8 5juo 
5JUO 5jup 
5JUP 5jus 
5JUS 5jut 
5JUT 5juu 
5JUU 5k0y 
5K0Y 5kcr 
5KCR 5kcs 
5KCS 5kps 
5KPS 5kpv 
5KPV 5kpw 
5KPW 5kpx 
5KPX 5l3p 
5L3P 5lmn 
5LMN 5lmo 
5LMO 5lmp 
5LMP 5lmq 
5LMQ 5lmr 
5LMR 5lms 
5LMS 5lmt 
5LMT 5lmu 
5LMU 5lmv 
5LMV 5lyb 
5LYB 5lza 
5LZA 5lzb 
5LZB 5lzc 
5LZC 5lzd 
5LZD 5lze 
5LZE 5lzf 
5LZF 5lzs 
5LZS 5lzt 
5LZT 5lzu 
5LZU 5lzv 
5LZV 5lzw 
5LZW 5lzx 
5LZX 5lzy 
5LZY 5lzz 
5LZZ 5tga 
5TGA - Links (links to other resources describing this domain)
-
PFAM S4 INTERPRO IPR002942 PROSITE RIBOSOMAL_S4

