The domain within your query sequence starts at position 938 and ends at position 961; the E-value for the ZnF_C2H2 domain shown below is 9.44e-2.
HQCRECGLCYTSHGSLARHLFIVH
ZnF_C2H2zinc finger |
|---|
| SMART accession number: | SM00355 |
|---|---|
| Description: | - |
| Interpro abstract (IPR013087): | C2H2-type (classical) zinc fingers (Znf) were the first class to be characterised. They contain a short beta hairpin and an alpha helix (beta/beta/alpha structure), where a single zinc atom is held in place by Cys(2)His(2) (C2H2) residues in a tetrahedral array. C2H2 Znf's can be divided into three groups based on the number and pattern of fingers: triple-C2H2 (binds single ligand), multiple-adjacent-C2H2 (binds multiple ligands), and separated paired-C2H2 [ (PUBMED:11361095) ]. C2H2 Znf's are the most common DNA-binding motifs found in eukaryotic transcription factors, and have also been identified in prokaryotes [ (PUBMED:10664601) ]. Transcription factors usually contain several Znf's (each with a conserved beta/beta/alpha structure) capable of making multiple contacts along the DNA, where the C2H2 Znf motifs recognise DNA sequences by binding to the major groove of DNA via a short alpha-helix in the Znf, the Znf spanning 3-4 bases of the DNA [ (PUBMED:10940247) ]. C2H2 Znf's can also bind to RNA and protein targets [ (PUBMED:18253864) ]. Zinc finger (Znf) domains are relatively small protein motifs which contain multiple finger-like protrusions that make tandem contacts with their target molecule. Some of these domains bind zinc, but many do not; instead binding other metals such as iron, or no metal at all. For example, some family members form salt bridges to stabilise the finger-like folds. They were first identified as a DNA-binding motif in transcription factor TFIIIA from Xenopus laevis (African clawed frog), however they are now recognised to bind DNA, RNA, protein and/or lipid substrates [ (PUBMED:10529348) (PUBMED:15963892) (PUBMED:15718139) (PUBMED:17210253) (PUBMED:12665246) ]. Their binding properties depend on the amino acid sequence of the finger domains and of the linker between fingers, as well as on the higher-order structures and the number of fingers. Znf domains are often found in clusters, where fingers can have different binding specificities. There are many superfamilies of Znf motifs, varying in both sequence and structure. They display considerable versatility in binding modes, even between members of the same class (e.g. some bind DNA, others protein), suggesting that Znf motifs are stable scaffolds that have evolved specialised functions. For example, Znf-containing proteins function in gene transcription, translation, mRNA trafficking, cytoskeleton organisation, epithelial development, cell adhesion, protein folding, chromatin remodelling and zinc sensing, to name but a few [ (PUBMED:11179890) ]. Zinc-binding motifs are stable structures, and they rarely undergo conformational changes upon binding their target. This entry represents the classical C2H2 zinc finger domain. |
| Family alignment: |
There are 2594488 ZnF_C2H2 domains in 441262 proteins in SMART's nrdb database.
Click on the following links for more information.
- Evolution (species in which this domain is found)
-
Taxonomic distribution of proteins containing ZnF_C2H2 domain.
This tree includes only several representative species. The complete taxonomic breakdown of all proteins with ZnF_C2H2 domain is also avaliable.
Click on the protein counts, or double click on taxonomic names to display all proteins containing ZnF_C2H2 domain in the selected taxonomic class.
- Literature (relevant references for this domain)
-
Primary literature is listed below; Automatically-derived, secondary literature is also avaliable.
- Parraga G et al.
- Zinc-dependent structure of a single-finger domain of yeast ADR1.
- Science. 1988; 241: 1489-92
- Display abstract
In the proposed "zinc finger" DNA-binding motif, each repeat unit binds a zinc metal ion through invariant Cys and His residues and this drives the folding of each 30-residue unit into an independent nucleic acid-binding domain. To obtain structural information, we synthesized single and double zinc finger peptides from the yeast transcription activator ADR1, and assessed the metal-binding and DNA-binding properties of these peptides, as well as the solution structure of the metal-stabilized domains, with the use of a variety of spectroscopic techniques. A single zinc finger can exist as an independent structure sufficient for zinc-dependent DNA binding. An experimentally determined model of the single finger is proposed that is consistent with circular dichroism, one- and two-dimensional nuclear magnetic resonance, and visual spectroscopy of the single-finger peptide reconstituted in the presence of zinc.
- Disease (disease genes where sequence variants are found in this domain)
-
SwissProt sequences and OMIM curated human diseases associated with missense mutations within the ZnF_C2H2 domain.
Protein Disease Wilms tumor protein (P19544) (SMART) OMIM:194070: Wilms tumor, type 1 ; Denys-Drash syndrome ; Frasier syndrome
OMIM:136680:Transcriptional activator GLI3 (P10071) (SMART) OMIM:165240: Greig cephalopolysyndactyly syndrome
OMIM:175700: Pallister-Hall syndrome
OMIM:146510: Polydactyly, preaxial, type IV
OMIM:174700: Polydactyly, postaxial, types A1 and B
OMIM:174200: - Metabolism (metabolic pathways involving proteins which contain this domain)
-
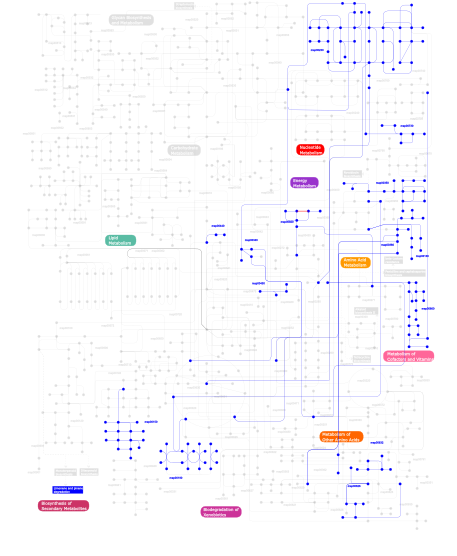
Click the image to view the interactive version of the map in iPath% proteins involved KEGG pathway ID Description 15.88 map04340 Hedgehog signaling pathway 12.94 map05217 Basal cell carcinoma 8.24 map04520 Adherens junction 7.06 map04010 MAPK signaling pathway 4.71 map05220 Chronic myeloid leukemia 4.71 map00908 Zeatin biosynthesis 4.12 map04350 TGF-beta signaling pathway 4.12 map05221 Acute myeloid leukemia 4.12 map04115 p53 signaling pathway 3.53 map00903 Limonene and pinene degradation 3.53 map05215 Prostate cancer 3.53  map00632
map00632Benzoate degradation via CoA ligation 2.35 map03050 Proteasome 2.35 map03022 Basal transcription factors 1.76  map00350
map00350Tyrosine metabolism 1.76  map00450
map00450Selenoamino acid metabolism 1.76  map00340
map00340Histidine metabolism 1.76  map00380
map00380Tryptophan metabolism 1.76  map00440
map00440Aminophosphonate metabolism 1.76  map00626
map00626Naphthalene and anthracene degradation 1.76  map00150
map00150Androgen and estrogen metabolism 1.18  map00190
map00190Oxidative phosphorylation 1.18  map00130
map00130Ubiquinone biosynthesis 1.18 map04320 Dorso-ventral axis formation 1.18  map00920
map00920Sulfur metabolism 0.59  map00860
map00860Porphyrin and chlorophyll metabolism 0.59  map00730
map00730Thiamine metabolism 0.59  map00230
map00230Purine metabolism This information is based on mapping of SMART genomic protein database to KEGG orthologous groups. Percentage points are related to the number of proteins with ZnF_C2H2 domain which could be assigned to a KEGG orthologous group, and not all proteins containing ZnF_C2H2 domain. Please note that proteins can be included in multiple pathways, ie. the numbers above will not always add up to 100%.
- Structure (3D structures containing this domain)
3D Structures of ZnF_C2H2 domains in PDB
PDB code Main view Title 1a1f 
DSNR (ZIF268 VARIANT) ZINC FINGER-DNA COMPLEX (GACC SITE) 1a1g 
DSNR (ZIF268 VARIANT) ZINC FINGER-DNA COMPLEX (GCGT SITE) 1a1h 
QGSR (ZIF268 VARIANT) ZINC FINGER-DNA COMPLEX (GCAC SITE) 1a1i 
RADR (ZIF268 VARIANT) ZINC FINGER-DNA COMPLEX (GCAC SITE) 1a1j 
RADR (ZIF268 VARIANT) ZINC FINGER-DNA COMPLEX (GCGT SITE) 1a1k 
RADR (ZIF268 VARIANT) ZINC FINGER-DNA COMPLEX (GACC SITE) 1a1l 
ZIF268 ZINC FINGER-DNA COMPLEX (GCAC SITE) 1aay 
ZIF268 ZINC FINGER-DNA COMPLEX 1ard 
STRUCTURES OF DNA-BINDING MUTANT ZINC FINGER DOMAINS: IMPLICATIONS FOR DNA BINDING 1are 
STRUCTURES OF DNA-BINDING MUTANT ZINC FINGER DOMAINS: IMPLICATIONS FOR DNA BINDING 1arf 
STRUCTURES OF DNA-BINDING MUTANT ZINC FINGER DOMAINS: IMPLICATIONS FOR DNA BINDING 1bbo 
HIGH-RESOLUTION SOLUTION STRUCTURE OF THE DOUBLE CYS2*HIS2 ZINC FINGER FROM THE HUMAN ENHANCER BINDING PROTEIN MBP-1 1bhi 
STRUCTURE OF TRANSACTIVATION DOMAIN OF CRE-BP1/ATF-2, NMR, 20 STRUCTURES 1ej6 
Reovirus core 1f2i 
COCRYSTAL STRUCTURE OF SELECTED ZINC FINGER DIMER BOUND TO DNA 1fv5 
SOLUTION STRUCTURE OF THE FIRST ZINC FINGER FROM THE DROSOPHILA U-SHAPED TRANSCRIPTION FACTOR 1g2d 
STRUCTURE OF A CYS2HIS2 ZINC FINGER/TATA BOX COMPLEX (CLONE #2) 1g2f 
STRUCTURE OF A CYS2HIS2 ZINC FINGER/TATA BOX COMPLEX (TATAZF;CLONE #6) 1jk1 
Zif268 D20A Mutant Bound to WT DNA Site 1jk2 
Zif268 D20A mutant bound to the GCT DNA site 1jn7 
Solution Structure of a CCHH mutant of the ninth CCHC Zinc Finger of U-shaped 1klr 
NMR Structure of the ZFY-6T[Y10F] Zinc Finger 1kls 
NMR Structure of the ZFY-6T[Y10L] Zinc Finger 1llm 
Crystal Structure of a Zif23-GCN4 Chimera Bound to DNA 1mey 
CRYSTAL STRUCTURE OF A DESIGNED ZINC FINGER PROTEIN BOUND TO DNA 1ncs 
NMR STUDY OF SWI5 ZINC FINGER DOMAIN 1 1njq 
NMR structure of the single QALGGH zinc finger domain from Arabidopsis thaliana SUPERMAN protein 1p47 
Crystal Structure of tandem Zif268 molecules complexed to DNA 1p7a 
Solution Stucture of the Third Zinc Finger from BKLF 1paa 
STRUCTURE OF A HISTIDINE-X4-HISTIDINE ZINC FINGER DOMAIN: INSIGHTS INTO ADR1-UAS1 PROTEIN-DNA RECOGNITION 1rik 
E6-binding zinc finger (E6apc1) 1rim 
E6-binding zinc finger (E6apc2) 1rmd 
RAG1 DIMERIZATION DOMAIN 1sp1 
NMR STRUCTURE OF A ZINC FINGER DOMAIN FROM TRANSCRIPTION FACTOR SP1F3, MINIMIZED AVERAGE STRUCTURE 1sp2 
NMR STRUCTURE OF A ZINC FINGER DOMAIN FROM TRANSCRIPTION FACTOR SP1F2, MINIMIZED AVERAGE STRUCTURE 1srk 
Solution structure of the third zinc finger domain of FOG-1 1tf3 
TFIIIA FINGER 1-3 BOUND TO DNA, NMR, 22 STRUCTURES 1tf6 
CO-CRYSTAL STRUCTURE OF XENOPUS TFIIIA ZINC FINGER DOMAIN BOUND TO THE 5S RIBOSOMAL RNA GENE INTERNAL CONTROL REGION 1u85 
ARG326-TRP mutant of the third zinc finger of BKLF 1u86 
321-TW-322 insertion mutant of the third zinc finger of BKLF 1ubd 
CO-CRYSTAL STRUCTURE OF HUMAN YY1 ZINC FINGER DOMAIN BOUND TO THE ADENO-ASSOCIATED VIRUS P5 INITIATOR ELEMENT 1un6 
THE CRYSTAL STRUCTURE OF A ZINC FINGER - RNA COMPLEX REVEALS TWO MODES OF MOLECULAR RECOGNITION 1va1 
Solution Structure of Transcription Factor Sp1 DNA Binding Domain (Zinc Finger 1) 1va2 
Solution Structure of Transcription Factor Sp1 DNA Binding Domain (Zinc Finger 2) 1va3 
Solution Structure of Transcription Factor Sp1 DNA Binding Domain (Zinc Finger 3) 1wir 
Solution structure of the C2H2 zinc finger domain of the protein arginine N-methyltransferase 3 from Mus musculus 1wjp 
Solution structure of zf-C2H2 domains from human Zinc finger protein 295 1x3c 
Solution structure of the C2H2 type zinc-binding domain of human zinc finger protein 292 1x5w 
Solution structure of the C2H2 type zinc-binding domain of human zinc finger protein 64, isoforms 1 and 2 1x6e 
Solution structures of the C2H2 type zinc finger domain of human Zinc finger protein 24 1x6f 
Solution structures of the C2H2 type zinc finger domain of human Zinc finger protein 462 1x6h 
Solution structures of the C2H2 type zinc finger domain of human Transcriptional repressor CTCF 1xf7 
High Resolution NMR Structure of the Wilms' Tumor Suppressor Protein (WT1) Finger 3 1xrz 
NMR Structure of a Zinc Finger with Cyclohexanylalanine Substituted for the Central Aromatic Residue 1y0j 
Zinc fingers as protein recognition motifs: structural basis for the GATA-1/Friend of GATA interaction 1yui 
SOLUTION NMR STRUCTURE OF THE GAGA FACTOR/DNA COMPLEX, REGULARIZED MEAN STRUCTURE 1yuj 
SOLUTION NMR STRUCTURE OF THE GAGA FACTOR/DNA COMPLEX, 50 STRUCTURES 1zaa 
ZINC FINGER-DNA RECOGNITION: CRYSTAL STRUCTURE OF A ZIF268-DNA COMPLEX AT 2.1 ANGSTROMS 1zfd 
SWI5 ZINC FINGER DOMAIN 2, NMR, 45 STRUCTURES 1znf 
THREE-DIMENSIONAL SOLUTION STRUCTURE OF A SINGLE ZINC FINGER DNA-BINDING DOMAIN 1znm 
A ZINC FINGER WITH AN ARTIFICIAL BETA-TURN, ORIGINAL SEQUENCE TAKEN FROM THE THIRD ZINC FINGER DOMAIN OF THE HUMAN TRANSCRIPTIONAL REPRESSOR PROTEIN YY1 (YING AND YANG 1, A DELTA TRANSCRIPTION FACTOR), NMR, 34 STRUCTURES 1zr9 
Solution Structure of a Human C2H2-type Zinc Finger Protein 1zu1 
Solution Structure of the N-terminal Zinc Fingers of the Xenopus laevis double stranded RNA binding protein ZFa 1zw8 
Solution structure of a ZAP1 zinc-responsive domain provides insights into metalloregulatory transcriptional repression in Saccharomyces cerevisiae 2ab3 
Solution structures and characterization of HIV RRE IIB RNA targeting zinc finger proteins 2ab7 
Solution structures and characterization of HIV RRE IIB RNA targeting zinc finger proteins 2adr 
ADR1 DNA-BINDING DOMAIN FROM SACCHAROMYCES CEREVISIAE, NMR, 25 STRUCTURES 2cot 
Solution structure of the first and second zf-C2H2 domain of Zinc finger protein 435 2cse 
Features of Reovirus Outer-Capsid Protein mu1 Revealed by Electron and Image Reconstruction of the virion at 7.0-A Resolution 2csh 
Solution structure of tandem repeat of the zf-C2H2 domains of human zinc finger protein 297B 2ct1 
Solution Structure of the zinc finger domain of Transcriptional repressor CTCF protein 2ctd 
Solution structure of two zf-C2H2 domains from human Zinc finger protein 512 2czr 
Crystal structure of TBP-interacting protein (Tk-TIP26) and implications for its inhibition mechanism of the interaction between TBP and TATA-DNA 2d9h 
Solution structure of the forth and fifth zf-C2H2 domains of zinc finger protein 692 2dlk 
Solution structure of the first and the second zf-C2H2 domains of zinc finger protein 692 2dlq 
Solution structure of the tandem four zf-C2H2 domain repeats of murine GLI-Kruppel family member HKR3 2dmd 
Solution structure of the N-terminal C2H2 type zinc-binding domain of the Zinc finger protein 64, isoforms 1 and 2 2dmi 
Solution structure of the first and the second zf-C2H2 like domains of human Teashirt homolog 3 2drp 
THE CRYSTAL STRUCTURE OF A TWO ZINC-FINGER PEPTIDE REVEALS AN EXTENSION TO THE RULES FOR ZINC-FINGER/DNA RECOGNITION 2ebt 
Solution structure of three tandem repeats of zf-C2H2 domains from human Kruppel-like factor 5 2ee8 
Solution structure of three zf-C2H2 domains from mouse protein odd-skipped-related 2 splicing isoform 2 2ej4 
Functional and structural basis of nuclear localization signal in ZIC3 zinc finger domain: a role of conserved tryptophan residue in the zinc finger domain 2el4 
Solution structure of the 15th zf-C2H2 domain from human Zinc finger protein 268 2el5 
Solution structure of the 18th zf-C2H2 domain from human Zinc finger protein 268 2el6 
Solution structure of the 21th zf-C2H2 domain from human Zinc finger protein 268 2elm 
Solution structure of the 10th C2H2 zinc finger of human Zinc finger protein 406 2eln 
Solution structure of the 11th C2H2 zinc finger of human Zinc finger protein 406 2elo 
Solution structure of the 12th C2H2 zinc finger of human Zinc finger protein 406 2elq 
Solution structure of the 14th C2H2 zinc finger of human Zinc finger protein 406 2elr 
Solution structure of the 15th C2H2 zinc finger of human Zinc finger protein 406 2els 
Solution structure of the 2nd C2H2 zinc finger of human Zinc finger protein 406 2elt 
Solution structure of the 3rd C2H2 zinc finger of human Zinc finger protein 406 2elu 
Solution structure of the 5th C2H2 zinc finger of human Zinc finger protein 406 2elv 
Solution structure of the 6th C2H2 zinc finger of human Zinc finger protein 406 2elw 
Solution structure of the 5th C2H2 zinc finger of mouse Zinc finger protein 406 2elx 
Solution structure of the 8th C2H2 zinc finger of mouse Zinc finger protein 406 2ely 
Solution structure of the third zf-C2H2 domain from human Zinc finger protein 224 2elz 
Solution structure of the 17th zf-C2H2 domain from human Zinc finger protein 224 2em0 
Solution structure of the 18th zf-C2H2 domain from human Zinc finger protein 224 2em1 
Solution structure of the C2H2 type zinc finger (region 637-667) of human Zinc finger protein 268 2em2 
Solution structure of the C2H2 type zinc finger (region 584-616) of human Zinc finger protein 28 homolog 2em3 
Solution structure of the C2H2 type zinc finger (region 640-672) of human Zinc finger protein 28 homolog 2em4 
Solution structure of the C2H2 type zinc finger (region 724-756) of human Zinc finger protein 28 homolog 2em5 
Solution structure of the C2H2 type zinc finger (region 768-800) of human Zinc finger protein 95 homolog 2em6 
Solution structure of the C2H2 type zinc finger (region 199-231) of human Zinc finger protein 224 2em7 
Solution structure of the C2H2 type zinc finger (region 339-371) of human Zinc finger protein 224 2em8 
Solution structure of the C2H2 type zinc finger (region 423-455) of human Zinc finger protein 224 2em9 
Solution structure of the C2H2 type zinc finger (region 367-399) of human Zinc finger protein 224 2ema 
Solution structure of the C2H2 type zinc finger (region 312-344) of human Zinc finger protein 347 2emb 
Solution structure of the C2H2 type zinc finger (region 342-372) of human Zinc finger protein 473 2emc 
Solution structure of the C2H2 type zinc finger (region 641-673) of human Zinc finger protein 473 2eme 
Solution structure of the C2H2 type zinc finger (region 725-757) of human Zinc finger protein 473 2emf 
Solution structure of the C2H2 type zinc finger (region 379-411) of human Zinc finger protein 484 2emg 
Solution structure of the C2H2 type zinc finger (region 463-495) of human Zinc finger protein 484 2emh 
Solution structure of the C2H2 type zinc finger (region 491-523) of human Zinc finger protein 484 2emi 
Solution structure of the C2H2 type zinc finger (region 547-579) of human Zinc finger protein 484 2emj 
Solution structure of the C2H2 type zinc finger (region 612-644) of human Zinc finger protein 28 homolog 2emk 
Solution structure of the C2H2 type zinc finger (region 668-700) of human Zinc finger protein 28 homolog 2eml 
Solution structure of the C2H2 type zinc finger (region 752-784) of human Zinc finger protein 28 homolog 2emm 
Solution structure of the C2H2 type zinc finger (region 544-576) of human Zinc finger protein 95 homolog 2emp 
Solution structure of the C2H2 type zinc finger (region 536-568) of human Zinc finger protein 347 2emv 
Solution structure of the C2H2 type zinc finger (region 859-889) of human Zinc finger protein 268 2emw 
Solution structure of the C2H2 type zinc finger (region 301-331) of human Zinc finger protein 268 2emx 
Solution structure of the C2H2 type zinc finger (region 273-303) of human Zinc finger protein 268 2emy 
Solution structure of the C2H2 type zinc finger (region 551-583) of human Zinc finger protein 268 2emz 
Solution structure of the C2H2 type zinc finger (region 628-660) of human Zinc finger protein 95 homolog 2en0 
Solution structure of the C2H2 type zinc finger (region 385-413) of human Zinc finger protein 268 2en1 
Solution structure of the C2H2 type zinc finger (region 563-595) of human Zinc finger protein 224 2en2 
Solution structure of the C2H2 type zinc finger (region 598-626) of human B-cell lymphoma 6 protein 2en3 
Solution structure of the C2H2 type zinc finger (region 796-828) of human Zinc finger protein 95 homolog 2en4 
Solution structure of the C2H2 type zinc finger (region 284-316) of human Zinc finger protein 347 2en6 
Solution structure of the C2H2 type zinc finger (region 887-919) of human Zinc finger protein 268 2en7 
Solution structure of the C2H2 type zinc finger (region 495-525) of human Zinc finger protein 268 2en8 
Solution structure of the C2H2 type zinc finger (region 171-203) of human Zinc finger protein 224 2en9 
Solution structure of the C2H2 type zinc finger (region 415-447) of human Zinc finger protein 28 homolog 2ena 
Solution structure of the C2H2 type zinc finger (region 311-343) of human Zinc finger protein 224 2enc 
Solution structure of the C2H2 type zinc finger (region 395-427) of human Zinc finger protein 224 2ene 
Solution structure of the C2H2 type zinc finger (region 592-624) of human Zinc finger protein 347 2enf 
Solution structure of the C2H2 type zinc finger (region 340-372) of human Zinc finger protein 347 2enh 
Solution structure of the C2H2 type zinc finger (region 556-588) of human Zinc finger protein 28 homolog 2ent 
Solution structure of the second C2H2-type zinc finger domain from human Krueppel-like factor 15 2eoe 
Solution structure of the C2H2 type zinc finger (region 508-540) of human Zinc finger protein 347 2eof 
Solution structure of the C2H2 type zinc finger (region 411-441) of human Zinc finger protein 268 2eog 
Solution structure of the C2H2 type zinc finger (region 693-723) of human Zinc finger protein 268 2eoh 
Solution structure of the C2H2 type zinc finger (region 780-812) of human Zinc finger protein 28 homolog 2eoi 
Solution structure of the C2H2 type zinc finger (region 329-359) of human Zinc finger protein 268 2eoj 
Solution structure of the C2H2 type zinc finger (region 355-385) of human Zinc finger protein 268 2eok 
Solution structure of the C2H2 type zinc finger (region 441-469) of human Zinc finger protein 268 2eol 
Solution structure of the C2H2 type zinc finger (region 581-609) of human Zinc finger protein 268 2eom 
Solution structure of the C2H2 type zinc finger (region 341-373) of human Zinc finger protein 95 homolog 2eon 
Solution structure of the C2H2 type zinc finger (region 397-429) of human Zinc finger protein 95 homolog 2eoo 
Solution structure of the C2H2 type zinc finger (region 425-457) of human Zinc finger protein 95 homolog 2eop 
Solution structure of the C2H2 type zinc finger (region 719-751) of human Zinc finger protein 268 2eoq 
Solution structure of the C2H2 type zinc finger (region 283-315) of human Zinc finger protein 224 2eor 
Solution structure of the C2H2 type zinc finger (region 255-287) of human Zinc finger protein 224 2eos 
Solution structure of the C2H2 type zinc finger (region 626-654) of human B-cell lymphoma 6 protein 2eou 
Solution structure of the C2H2 type zinc finger (region 370-400) of human Zinc finger protein 473 2eov 
Solution structure of the C2H2 type zinc finger (region 519-551) of human Zinc finger protein 484 2eow 
Solution structure of the C2H2 type zinc finger (region 368-400) of human Zinc finger protein 347 2eox 
Solution structure of the C2H2 type zinc finger (region 315-345) of human Zinc finger protein 473 2eoy 
Solution structure of the C2H2 type zinc finger (region 557-589) of human Zinc finger protein 473 2eoz 
Solution structure of the C2H2 type zinc finger (region 809-841) of human Zinc finger protein 473 2ep0 
Solution structure of the C2H2 type zinc finger (region 528-560) of human Zinc finger protein 28 homolog 2ep1 
Solution structure of the C2H2 type zinc finger (region 435-467) of human Zinc finger protein 484 2ep2 
Solution structure of the C2H2 type zinc finger (region 603-635) of human Zinc finger protein 484 2ep3 
Solution structure of the C2H2 type zinc finger (region 631-663) of human Zinc finger protein 484 2epa 
Solution structure of the first and second zf-C2H2 domains from human Krueppel-like factor 10 2epc 
Solution structure of Zinc finger domain 7 in Zinc finger protein 32 2epp 
Solution structure of the first C2H2 type zinc finger domain of Zinc finger protein 278 2epq 
Solution structure of the third zinc finger domain of Zinc finger protein 278 2epr 
Solution structure of the secound zinc finger domain of Zinc finger protein 278 2eps 
Solution structure of the 4th zinc finger domain of Zinc finger protein 278 2ept 
Solution structure of the first C2H2 type zinc finger domain of Zinc finger protein 32 2epu 
Solution structure of the secound C2H2 type zinc finger domain of Zinc finger protein 32 2epv 
Solution structure of the 20th C2H2 type zinc finger domain of Zinc finger protein 268 2epw 
Solution structure of the 24th C2H2 type zinc finger domain of Zinc finger protein 268 2epx 
Solution structure of the third C2H2 type zinc finger domain of Zinc finger protein 28 homolog 2epy 
Solution structure of the 10th C2H2 type zinc finger domain of Zinc finger protein 268 2epz 
Solution structure of the 4th C2H2 type zinc finger domain of Zinc finger protein 28 homolog 2eq0 
Solution structure of the 8th C2H2 type zinc finger domain of Zinc finger protein 347 2eq1 
Solution structure of the 9th C2H2 type zinc finger domain of Zinc finger protein 347 2eq2 
Solution structure of the 16th C2H2 type zinc finger domain of Zinc finger protein 347 2eq3 
Solution structure of the 17th C2H2 type zinc finger domain of Zinc finger protein 347 2eq4 
Solution structure of the 11th C2H2 type zinc finger domain of Zinc finger protein 224 2eqw 
Solution structure of the 6th C2H2 type zinc finger domain of Zinc finger protein 484 2ghf 
Solution structure of the complete zinc-finger region of human zinc-fingers and homeoboxes 1 (ZHX1) 2gli 
FIVE-FINGER GLI/DNA COMPLEX 2gqj 
Solution structure of the two zf-C2H2 like domains(493-575) of human zinc finger protein KIAA1196 2hgh 
Transcription Factor IIIA zinc fingers 4-6 bound to 5S rRNA 55mer (NMR structure) 2i13 
Aart, a six finger zinc finger designed to recognize ANN triplets 2j7j 
Invariance of the zinc finger module: a comparison of the free structure with those in nucleic-acid complexes 2jp9 
Structure of the Wilms Tumor Suppressor Protein Zinc Finger Domain Bound to DNA 2jpa 
Structure of the Wilms Tumor Suppressor Protein Zinc Finger Domain Bound to DNA 2jsp 
The prokaryotic Cys2His2 zinc finger adopts a novel fold as revealed by the NMR structure of A. tumefaciens Ros DNA binding domain 2kfq 
NMR Structure of FP1 2kmk 
Gfi-1 Zinc Fingers 3-5 complexed with DNA 2kvf 
Structure of the three-Cys2His2 domain of mouse testis zinc finger protein 2kvg 
Structure of the three-Cys2His2 domain of mouse testis zinc finger protein 2kvh 
Structure of the three-Cys2His2 domain of mouse testis zinc finger protein 2l1o 
Zinc to cadmium replacement in the A. thaliana SUPERMAN Cys2His2 zinc finger induces structural rearrangements of typical DNA base determinant positions 2l6z 
haddock model of GATA1NF:Lmo2LIM2-Ldb1LID with FOG 2lce 
Chemical shift assignment of Hr4436B from Homo Sapiens, Northeast Structural Genomics Consortium 2lt7 
Solution NMR structure of Kaiso zinc finger DNA binding domain in complex with Kaiso binding site DNA 2lv2 
Solution NMR structure of C2H2-type Zinc-fingers 4 and 5 from human Insulinoma-associated protein 1 (fragment 424-497), Northeast Structural Genomics Consortium Target HR7614B 2lvh 
Solution structure of the zinc finger AFV1p06 protein from the hyperthermophilic archaeal virus AFV1 2lvr 
Solution structure of Miz-1 zinc finger 8 2lvt 
Solution structure of Miz-1 zinc finger 9 2lvu 
Solution structure of Miz-1 zinc finger 10 2m0d 
Solution Structure of Miz-1 zinc finger 5 2m0e 
Solution Structure of Miz-1 zinc finger 6 2m0f 
Solution Structure of Miz-1 zinc finger 7 2m7q 
Solution structure of TAX1BP1 UBZ1+2 2m9a 
Solution NMR Structure of E3 ubiquitin-protein ligase ZFP91 from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR7784A 2ma7 
Solution NMR Structure of Zinc finger protein Eos from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR7992A 2mdg 
Solution NMR Structure of Zinc finger protein 423 from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR7298F 2mkd 
Human JAZ ZF3 Residues 168-227 2mkn 
Structural Characterization of Interactions between the Double-Stranded RNA-Binding Zinc Finger Protein JAZ and dsRNA 2mq1 
2MQ1 2n25 
2N25 2n26 
2N26 2nab 
2NAB 2prt 
Structure of the Wilms Tumor Suppressor Protein Zinc Finger Domain Bound to DNA 2rpc 
Solution structure of the tandem zf-C2H2 domains from the human zinc finger protein ZIC 3 2rsh 
Solution structures of the DNA-binding domains of immune-related zinc-finger protein ZFAT 2rsi 
Solution structures of the DNA-binding domains of immune-related zinc-finger protein ZFAT 2rsj 
Solution structures of the DNA-binding domains of immune-related zinc-finger protein ZFAT 2rut 
2RUT 2ruu 
2RUU 2ruv 
2RUV 2ruw 
2RUW 2rux 
2RUX 2ruy 
2RUY 2ruz 
2RUZ 2rv0 
2RV0 2rv2 
2RV2 2rv3 
2RV3 2rv4 
2RV4 2rv5 
2RV5 2rv6 
2RV6 2rv7 
2RV7 2wbs 
Crystal structure of the zinc finger domain of Klf4 bound to its target DNA 2wbt 
The Structure of a Double C2H2 Zinc Finger Protein from a Hyperthermophilic Archaeal Virus in the Absence of DNA 2wbu 
CRYSTAL STRUCTURE OF THE ZINC FINGER DOMAIN OF KLF4 BOUND TO ITS TARGET DNA 2yrh 
Solution structure of the C2H2-type zinc finger domain (699-729) from zinc finger protein 473 2yrj 
Solution structure of the C2H2-type zinc finger domain (781-813) from zinc finger protein 473 2yrk 
Solution structure of the zf-C2H2 domain in zinc finger homeodomain 4 2yrm 
Solution structure of the 1st zf-C2H2 domain from Human B-cell lymphoma 6 protein 2yso 
Solution structure of the C2H2 type zinc finger (region 656-688) of human Zinc finger protein 95 homolog 2ysp 
Solution structure of the C2H2 type zinc finger (region 507-539) of human Zinc finger protein 224 2ysv 
Solution structure of C2H2 type Zinc finger domain 17 in Zinc finger protein 473 2yt9 
Solution structure of C2H2 type Zinc finger domain 345 in Zinc finger protein 278 2yta 
Solution structure of C2H2 type Zinc finger domain 3 in Zinc finger protein 32 2ytb 
Solution structure of C2H2 type Zinc finger domain 5 in Zinc finger protein 32 2ytd 
Solution structure of the C2H2 type zinc finger (region 426-458) of human Zinc finger protein 473 2yte 
Solution structure of the C2H2 type zinc finger (region 484-512) of human Zinc finger protein 473 2ytf 
Solution structure of the C2H2 type zinc finger (region 607-639) of human Zinc finger protein 268 2ytg 
Solution structure of the C2H2 type zinc finger (region 369-401) of human Zinc finger protein 95 homolog 2yth 
Solution structure of the C2H2 type zinc finger (region 479-511) of human Zinc finger protein 224 2yti 
Solution structure of the C2H2 type zinc finger (region 564-596) of human Zinc finger protein 347 2ytj 
Solution structure of the C2H2 type zinc finger (region 771-803) of human Zinc finger protein 484 2ytk 
Solution structure of the C2H2 type zinc finger (region 396-428) of human Zinc finger protein 347 2ytm 
Solution structure of the C2H2 type zinc finger (region 696-728) of human Zinc finger protein 28 homolog 2ytn 
Solution structure of the C2H2 type zinc finger (region 732-764) of human Zinc finger protein 347 2yto 
Solution structure of the C2H2 type zinc finger (region 659-691) of human Zinc finger protein 484 2ytp 
Solution structure of the C2H2 type zinc finger (region 687-719) of human Zinc finger protein 484 2ytq 
Solution structure of the C2H2 type zinc finger (region 775-807) of human Zinc finger protein 268 2ytr 
Solution structure of the C2H2 type zinc finger (region 760-792) of human Zinc finger protein 347 2yts 
Solution structure of the C2H2 type zinc finger (region 715-747) of human Zinc finger protein 484 2ytt 
Solution structure of the C2H2 type zinc finger (region 204-236) of human Zinc finger protein 473 2yu5 
Solution structure of the zf-C2H2 domain (669-699AA) in zinc finger protein 473 2yu8 
Solution structure of the C2H2 type zinc finger (region 648-680) of human Zinc finger protein 347 3ax1 
Molecular insights into miRNA processing by Arabidopsis Serrate 3by4 
Structure of Ovarian Tumor (OTU) domain in complex with Ubiquitin 3c0r 
Structure of Ovarian Tumor (OTU) domain in complex with Ubiquitin 3iuf 
Crystal structure of the C2H2-type zinc finger domain of human ubi-d4 3iyl 
Atomic CryoEM Structure of a Nonenveloped Virus Suggests How Membrane Penetration Protein is Primed for Cell Entry 3jcr 
3JCR 3jct 
3JCT 3k1q 
Backbone model of an aquareovirus virion by cryo-electron microscopy and bioinformatics 3mjh 
Crystal Structure of Human Rab5A in complex with the C2H2 Zinc Finger of EEA1 3uk3 
Crystal structure of ZNF217 bound to DNA 3vk6 
Crystal structure of a phosphotyrosine binding domain 3w5k 
Crystal structure of Snail1 and importin beta complex 3zms 
LSD1-CoREST in complex with INSM1 peptide 3znf 
HIGH-RESOLUTION THREE-DIMENSIONAL STRUCTURE OF A SINGLE ZINC FINGER FROM A HUMAN ENHANCER BINDING PROTEIN IN SOLUTION 4bmj 
Structure of the UBZ1and2 tandem of the ubiquitin-binding adaptor protein TAX1BP1 4dgw 
Crystal Structure of the SF3a splicing factor complex of U2 snRNP 4f2j 
Crystal structure of ZNF217 bound to DNA, P6522 crystal form 4f6m 
Crystal structure of Kaiso zinc finger DNA binding domain in complex with Kaiso binding site DNA 4f6n 
Crystal structure of Kaiso zinc finger DNA binding protein in complex with methylated CpG site DNA 4gzn 
Mouse ZFP57 zinc fingers in complex with methylated DNA 4i1l 
Structural and Biological Features of FOXP3 Dimerization Relevant to Regulatory T Cell Function 4ijd 
Crystal structure of methyltransferase domain of human PR domain-containing protein 9 4is1 
Crystal structure of ZNF217 bound to DNA 4m9e 
Structure of Klf4 zinc finger DNA binding domain in complex with methylated DNA 4m9v 
Zfp57 mutant (E182Q) in complex with 5-carboxylcytosine DNA 4ndf 
Human Aprataxin (Aptx) bound to RNA-DNA, AMP, and Zn - product complex 4ndg 
Human Aprataxin (Aptx) bound to RNA-DNA and Zn - adenosine vanadate transition state mimic complex 4ndh 
Human Aprataxin (Aptx) bound to DNA, AMP, and Zn - product complex 4ndi 
Human Aprataxin (Aptx) AOA1 variant K197Q bound to RNA-DNA, AMP, and Zn - product complex 4r2a 
4R2A 4r2c 
4R2C 4r2d 
4R2D 4r2e 
4R2E 4r2p 
4R2P 4r2q 
4R2Q 4r2r 
4R2R 4r2s 
4R2S 4x9j 
4X9J 4z4k 
4Z4K 4znf 
HIGH-RESOLUTION THREE-DIMENSIONAL STRUCTURE OF A SINGLE ZINC FINGER FROM A HUMAN ENHANCER BINDING PROTEIN IN SOLUTION 5a7u 
5A7U 5aas 
5AAS 5apn 
5APN 5apo 
5APO 5dka 
5DKA 5egb 
5EGB 5eh2 
5EH2 5ei9 
5EI9 5gm6 
5GM6 5hzd 
5HZD 5i49 
5I49 5ido 
5IDO 5ion 
5ION 5ke6 
5KE6 5ke7 
5KE7 5ke8 
5KE8 5ke9 
5KE9 5kea 
5KEA 5keb 
5KEB 5kl2 
5KL2 5kl3 
5KL3 5kl4 
5KL4 5kl5 
5KL5 5kl6 
5KL6 5kl7 
5KL7 5znf 
ALTERNATING ZINC FINGERS IN THE HUMAN MALE ASSOCIATED PROTEIN ZFY: 2D NMR STRUCTURE OF AN EVEN FINGER AND IMPLICATIONS FOR "JUMPING-LINKER" DNA RECOGNITION 7znf 
ALTERNATING ZINC FINGERS IN THE HUMAN MALE ASSOCIATED PROTEIN ZFY: 2D NMR STRUCTURE OF AN EVEN FINGER AND IMPLICATIONS FOR "JUMPING-LINKER" DNA RECOGNITION - Links (links to other resources describing this domain)
-
PROSITE ZINC_FINGER_C2H2 PFAM zf-C2H2 INTERPRO IPR013087

